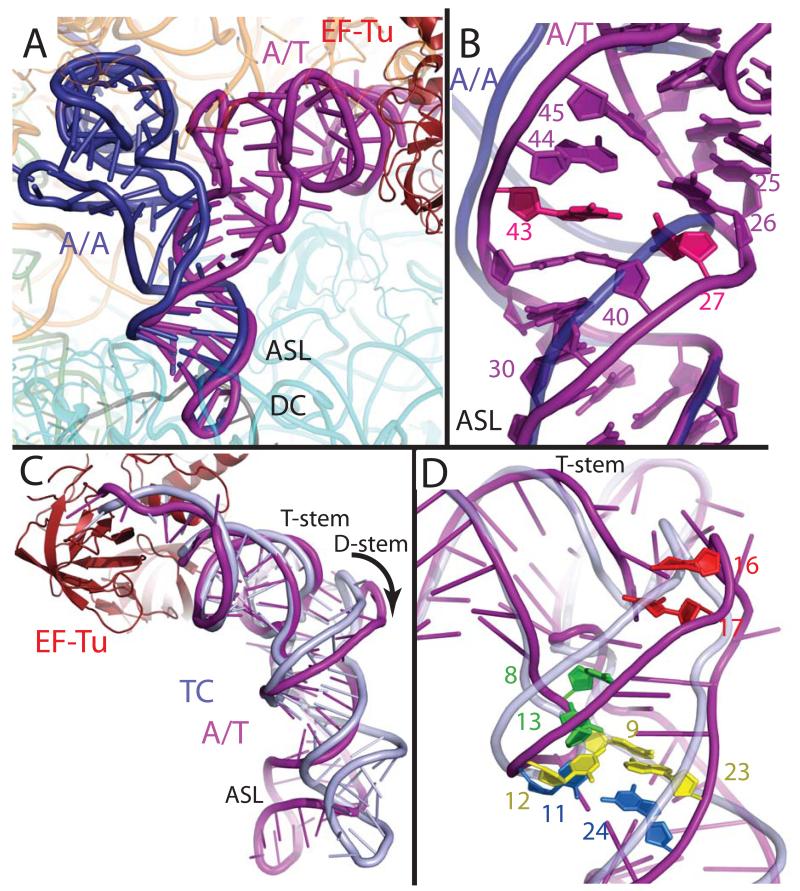
Fig. 2.
Distortion of aminoacyl-tRNA in the A/T state. (A) Comparison of the A/T tRNA (purple) with the fully accommodated canonical A/A tRNA (dark blue) (30) shows the overall extent of the tRNA disortion. (B) The structures diverge in the anticodon stem loop (ASL) with a reduction of helical twist after basepair 30:40, and separation of the phosphate-sugar backbones at nucleotides 25-45 and 26-44. Disruption of the 27:43 base pair (pink) (9) would facilitate this strand separation. (C) Comparison of the A/T tRNA with tRNA of the isolated TC (light blue) (22) highlights the swinging out and ~5Å shift of D-arm nucleotides. (D) The distortion rationalizes data pertaining to proflavin insertions at nucleotides 16-17 (bright red) (1), cross-linking of nucleotides 8 and 13 (green) (31), and mutations of the Hirsh base pair 24:11 (aqua) (6, 8) and 9:12:23 triple (yellow) (8).