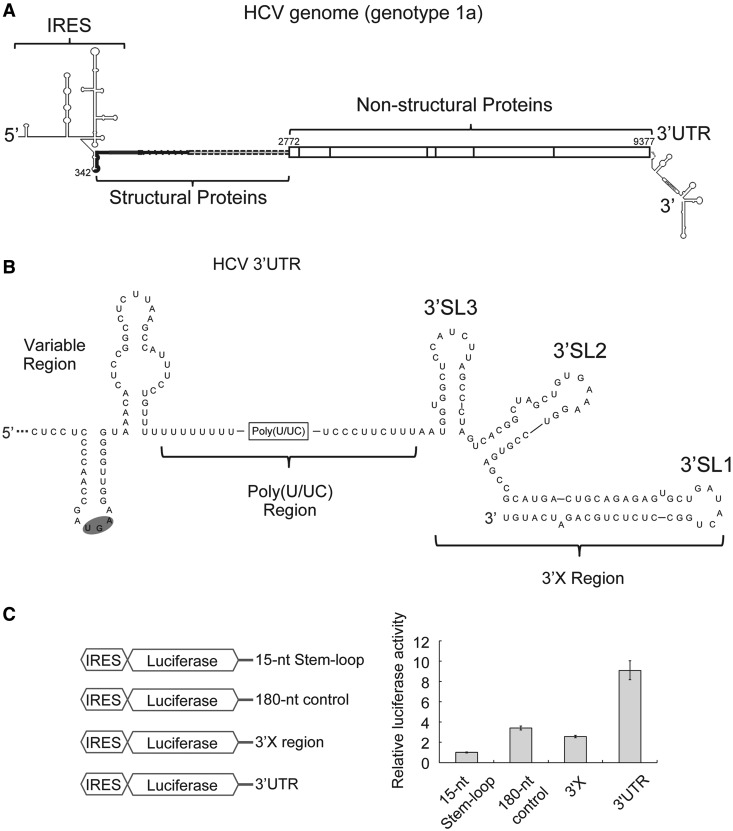
Figure 1.
The HCV 3′UTR stimulates IRES-dependent translation in cell culture. (A). Schematic drawing of the HCV genome. Secondary structures of both UTRs are indicated. The coding region is shown in thick lines for structural proteins and in boxes for non-structural proteins. Numbers are labeled according to genotype 1a strain H77 (B). Secondary structure of the 3′UTR of HCV genotype 1a. The variable region, poly(U/UC) region and three-stem loops in the 3′X region are labeled (C). Schematic drawing of different constructs used in translation assays are shown on the left. In the control construct, a 15-nt stem-loop (CUGCCGUAUAGGCAG) was attached to the 3′end the luciferase mRNA via a 5-nt linker (GUUCA) to ensure mRNA stability. The 180-nt control sequence is adopted from the pUC19 vector (447–630). On the right shows luciferase activities from cell-based translation assays using different RNA constructs. Luciferase activities in all experiments are normalized against that of the construct with a 15-nt stem loop downstream of the luciferase mRNA.