Figure 3.
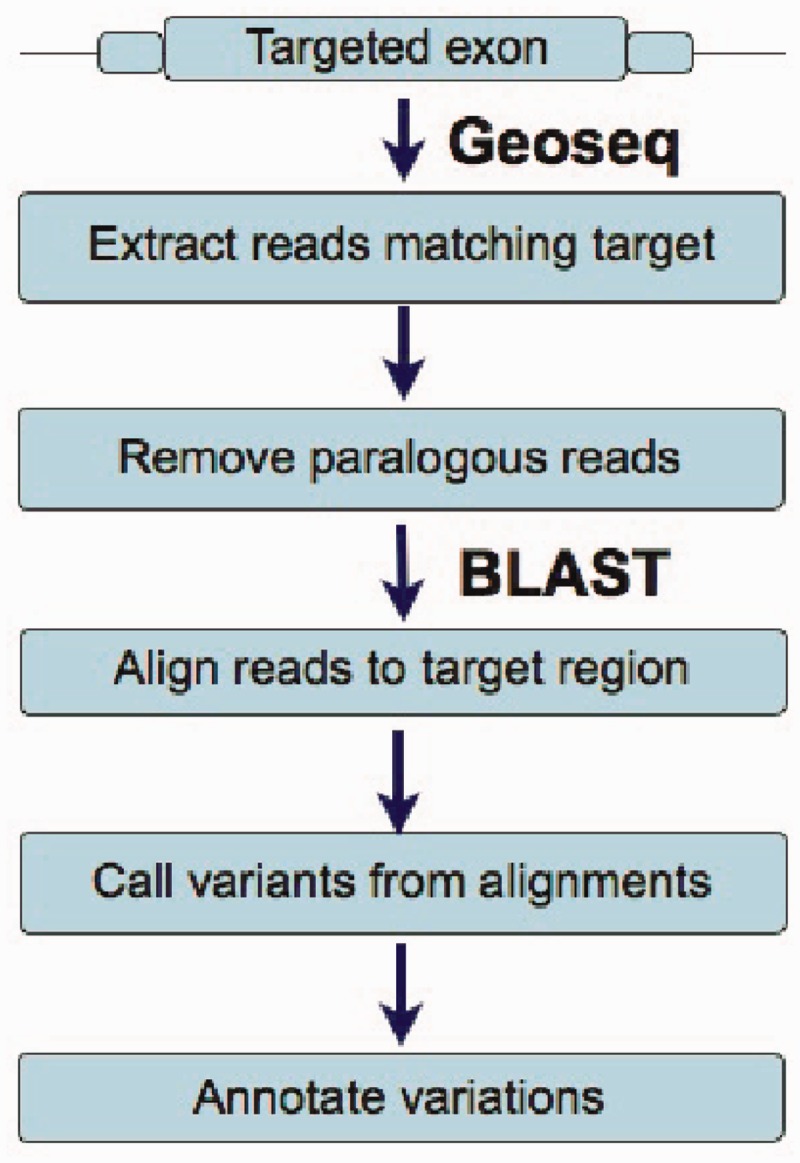
A schematic of the workflow used by MiST. The first step retrieves reads that can potentially match a targeted exonic region, and the following steps remove reads that can match other locations on the genome and align the reads to the exonic regions to call variants from the reference sequence.
