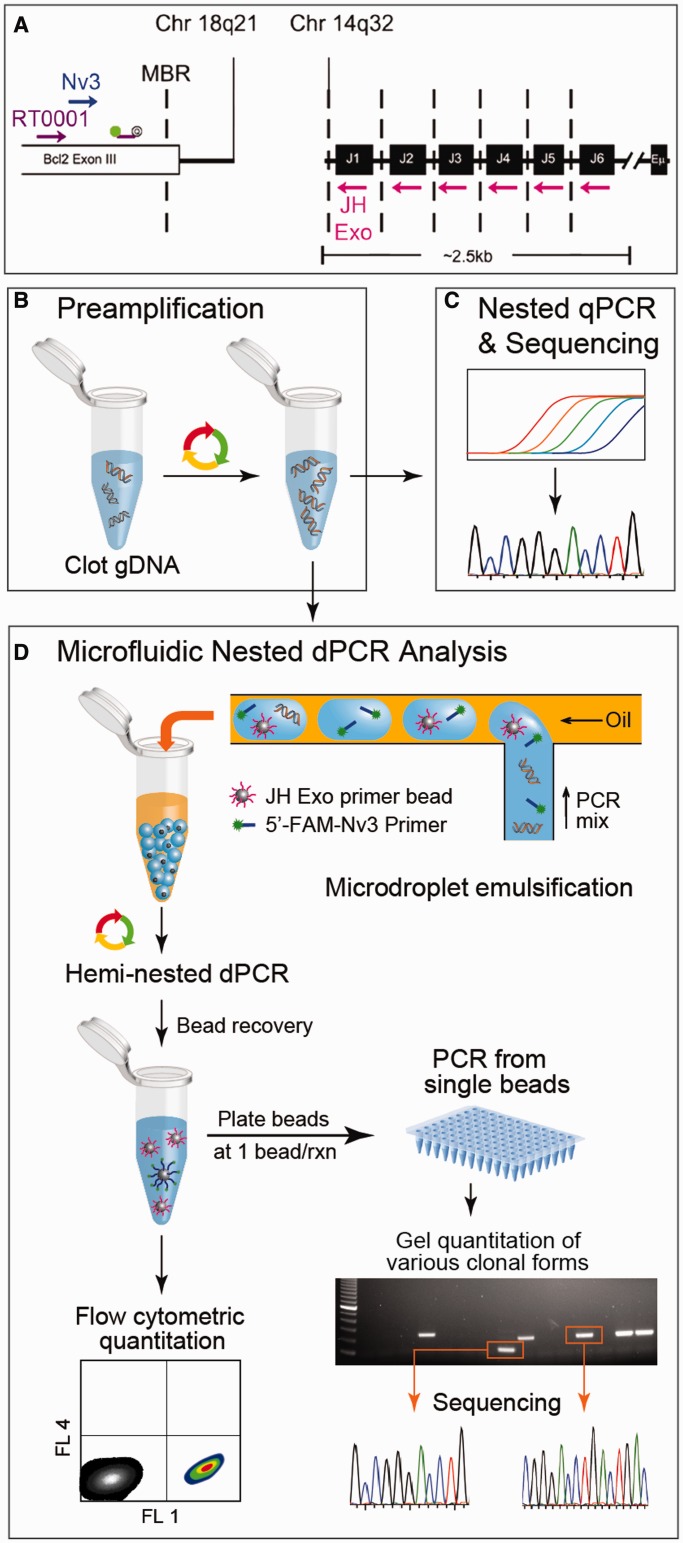
Figure 1.
Workflow and methodology for digital detection of t(14;18) in the clot gDNA of healthy subjects. (A) The PCR strategy for t(14;18) detection is illustrated by mapping the primers and probes used to their genomic targets. The first round of PCR was primed with the outer primer pair [consisting of an IgHJ consensus primer (JH Exo) and a BCL2-specific primer (RT0001)]. The hemi-nested secondary rounds of PCR were primed with a nested BCL2 primer (Nv3) while retaining the use of the IgHJ consensus primer (JH Exo). (B) A preamp PCR (20 cycles) produced t(14;18) amplicon from 3 µg of clot gDNA. The product of this first amplification step was then used as template in secondary rounds of hemi-nested PCR. Two different types of quantitative PCR were conducted: (C) standard qPCR using a BCL2-specific fluorescent probe (see Supplementary Figure S2); and (D) microfluidic emulsion single-molecule PCR using JH Exo-functionalized beads and 5′-FAM-labeled Nv3 for t(14;18) detection. A microfluidic emulsion generator array was used for high throughput (>106/h) production of monodisperse reaction volumes. Following emulsion generation, the nanoliter-scale reaction droplets were cycled to achieve single copy genetic analysis of the template. If a copy of t(14;18) and a bead were both present in a reaction droplet, then the bead was labeled with fluorescent amplicon during PCR, otherwise beads remained unlabeled. Beads were then recovered and analyzed by flow cytometry to determine t(14;18) concentration within a sample. In addition, some of the beads were distributed at ∼1 bead/well in 96-well plates to confirm that FAM+ concentration among beads corresponds with t(14;18)+ concentration as determined using a BCL2-specific probe. This tertiary round of amplification also produced sufficient template to conduct standard sequencing reactions that were derived from single molecule reactions.