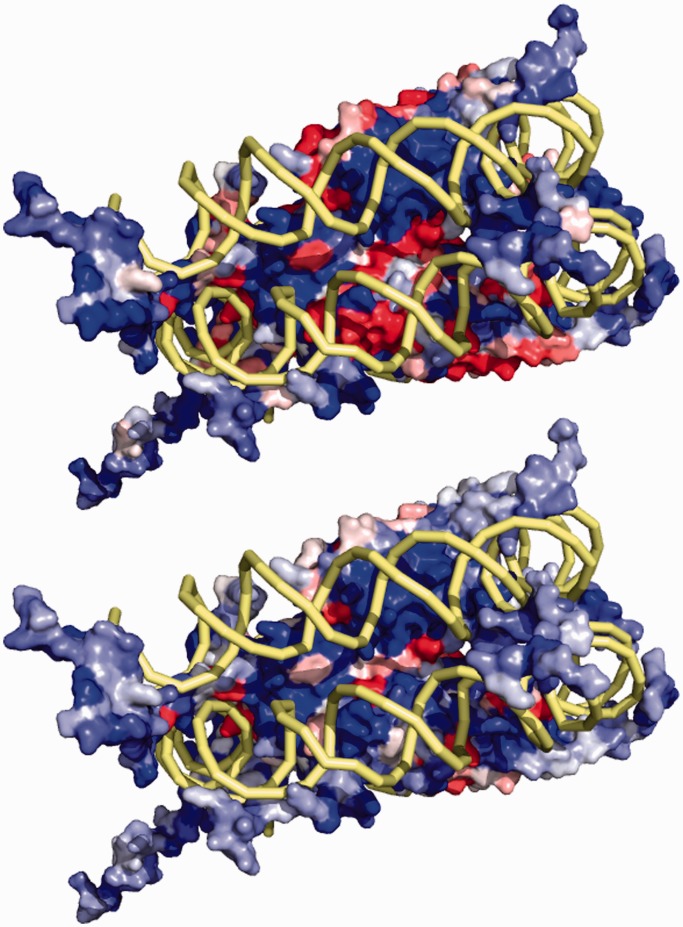
Figure 2.
The basic residue-level electrostatics feature is mapped onto the surface of the Nucleosome Core Particle (PDB 1KX5). The feature calculated in the shell between the van der Waals and solvent accessible surface (top) shows patches where this feature takes on negative values. When this feature is calculated for the shell that is shifted 0.5 Å outward, some patches flip from negative to positive. Thus, a region that might otherwise seem unfavorable to DNA binding is now seen to have the correct biophysical characteristics for recognition.