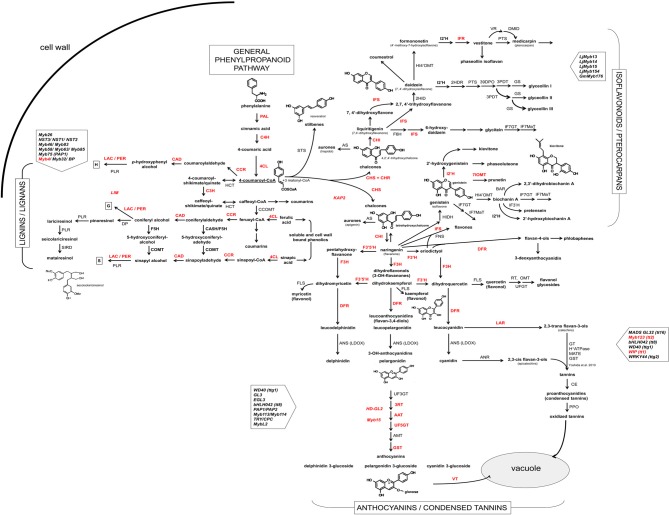
Figure 1.
Scheme of the phenylpropanoid pathway in common bean and soybean. Enzymes and transcription factors of the phenylpropanoid pathway [based on biochemical and genetic studies of several plant species and (KEGG Pathway database (http://www.genome.jp/kegg/pathway.html)] are presented in uppercase letters: PAL, phenylalanine ammonia lyase (EC:4.3.1.24); C4H, cinnamate 4-hydroxylase (EC:1.14.13.11); 4CL, 4-coumarate:CoA ligase (EC:6.2.1.12); C3H, 4-coumarate 3-hydroxylase (EC:1.14.13.-); COMT, caffeate O-methyltransferase (EC:2.1.1.68); F5H, ferulic acid 5-hydroxylase (EC:1.14.-.-); CCoOMT, caffeoyl CoA O-methyltransferase (EC:2.1.1.104); CCR, cinnamoyl CoA-reductase (EC:1.2.1.44); HCT, hydroxycinnamoyl-CoA: shikimate/guinate hydroxycinnamoyltransferase (EC:2.3.1.133); CAD, cinnamyl alcohol dehydrogenase (EC:1.1.1.195); LAC, laccase (EC:1.10.3.2); PER, lignin peroxydase (EC:1.11.1.7); PLR, pinoresinol/lariciresinol reductase; DP, dirigent protein; STS, stilbene synthase; CHS, chalcone synthase (EC:2.3.1.74); CHI, chalcone isomerase (EC:5.5.1.6); FNS, flavone synthase (EC:1.14.11.22); F3H, flavanone 3-hydroxylase (EC:1.14.11.9); F3′H, flavonoid 3′-hydroxylase (EC:1.14.13.21); F3′5′H, flavonoid 3′5′-hydroxylase (EC:1.14.13.88); FLS, flavonol synthase (EC:1.14.11.23); DFR, dihydroflavanol 4-reductase (EC:1.1.1.219); ANS (LDOX), anthocyanidin synthase (EC:1.14.11.19); UF3GT, UDP glucose flavonoid 3-O-glucosyltransferase (EC:2.4.1.115); 3RT, anthocyanidin-3-glucoside rhamnosyl transferase (EC:2.4.1.-); AAT, anthocyanin 5-aromatic acyltransferase (EC:2.3.1.153); AMT, anthocyanin methyltransferase; GST, glutathione S-transferase (EC:2.5.1.18); VT, vacuolar transporter; LAR, leucoanthocyanidin reductase (EC:1.17.1.3); ANR, anthocyanidin 4-reductase (1.3.1.77); GT, glucosyltransferase; MATE, multidrug and toxic compound extrusion; CE, condensed enzymes; PPO, polyphenol oxidase (EC:1.10.3.1); CHR, chalcone reductase (EC:1.1.1.-); IFS, 2-hydroxyisoflavanone synthase (EC:1.14.13.86); HIDH, 2-hydroxyisoflavanone dehydratase (EC:4.2.1.105); 7IOMT, isoflavone 7-O-methyltransferase (EC:2.1.1.150); I2'H, isoflavone 2′-hydroxylase (EC:1.14.13.89); IFR, isoflavanone reductase (EC:1.3.1.45); F6H, flavonoid 6-hydroxylase (EC:1.14.13.-); HI4′OMT, isoflavone 4-O-methyltransferase (EC:2.1.1.46); IF7MaT, isoflavone 7-O-methyltransferase (EC:2.3.1.115); VR, vestitone reductase (EC:1.1.1.-); DMID, dihydroxy-methoxy-isoflavanol dehydratase; G4DT, pterocarpan 4-dimethylallyltransferase; PTS, pterocarpan synthase (EC:1.1.1.246); KAP2, H-box-binding TF (CHS); Myb4, R2R3-type Myb TF (PAL); LIM1, TF LIM1 domain protein (lignin); HD-GL2, TF homeodomain protein GL2 like 1/ANL2; Myb15, R2R3-type Myb TF; WIP, TF zinc finger WIP (TT1); TGA1, basic domain/leucine zipper TF. Common bean genes cloned in this study are shown in bold red.