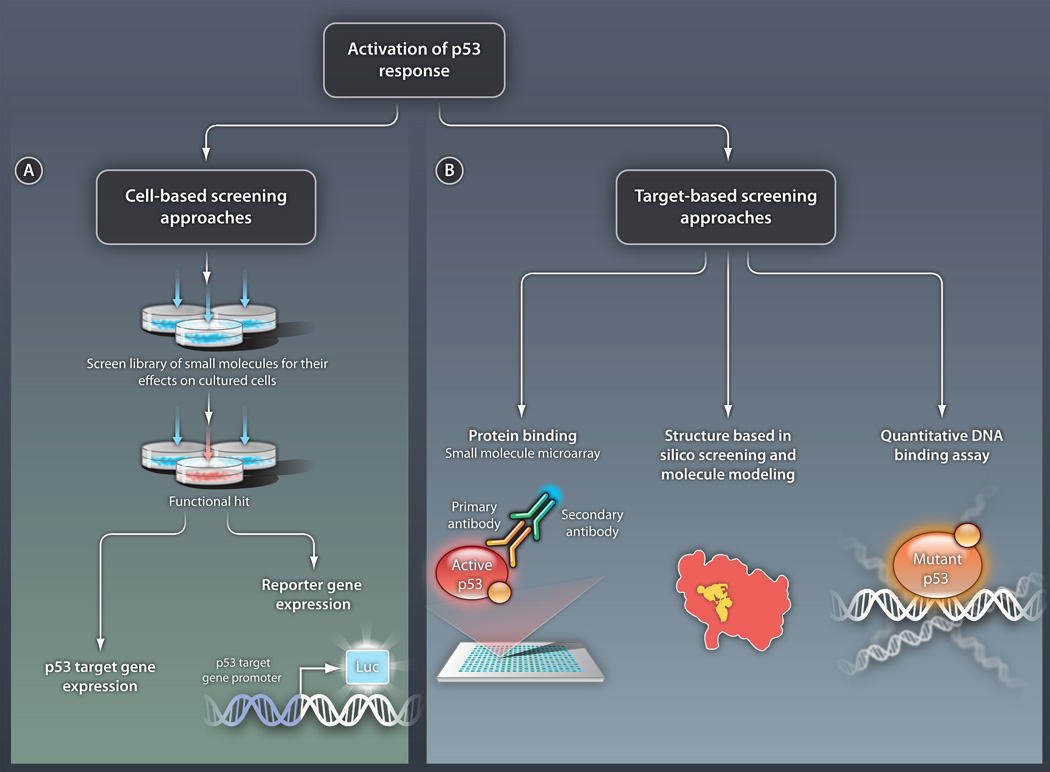
Fig 2. Strategies for identifying activators of the p53 response.
(A) Cell-based screening. In this approach, small molecules are tested for their ability to activate p53 pathways in cultured cells. Activation can be detected by measuring endogenous levels of transcripts from p53 target genes or by using various reporter systems. In this second method, the promoter of a p53 target gene is placed upstream of a gene encoding a product that can be easily monitored, such as the enzyme luciferase (luc). Cell-based screening will identify compounds with certain biological activity but with an unknown direct target molecule. (B) Target-based screening. In this approach, p53 protein activators are sought in vitro. Specific methods include the following: (i) Direct protein binding assays, such as the SMM assay. In this example, a library of small molecules is assembled on a microarray and the ability of p53 to bind the molecules is tested. Here, p53 binding is detected by a set of antibodies. (ii) Forward structural design, in which the structures of mutant versions of p53 are analyzed and molecules are designed to interact with relevant regions of these structures. (iii) Direct DNA binding assays, in which molecules that allow mutant p53 to bind DNA are sought. Such methods will enable the identification of compounds with a defined molecular mechanism.
CREDIT: C. BICKEL/SCIENCE TRANSLATIONAL MEDICINE