Figure 1.
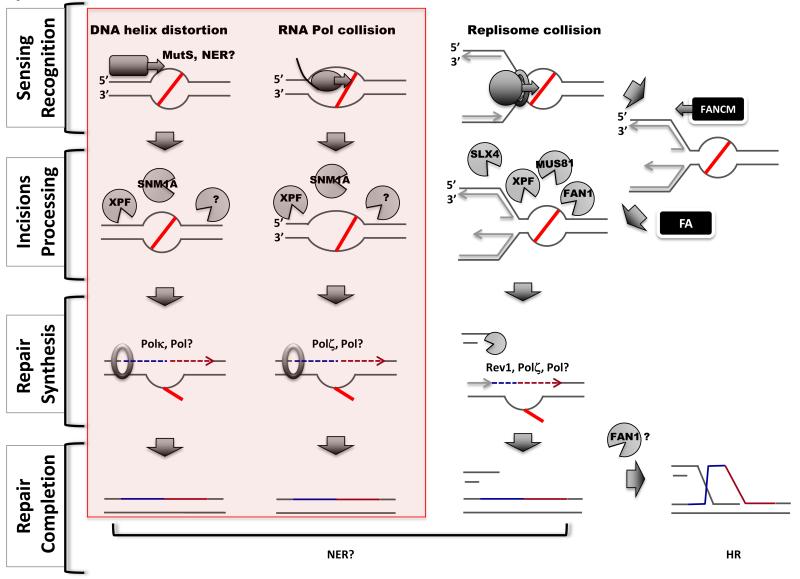
Pathways of ICL repair. Replication-independent mechanisms are highlighted in the pink box (left). Four steps are shown (left). Three distinct mechanisms can sense ICL lesions (indicated by a red line) (top): repair proteins recognizing distortion in the DNA helix (left column), a transcribing polymerase (middle column) or a replisome blocked by the lesion (right column). Recognition can occur through dedicated repair enzymes (NER or MMR) or following collision during DNA transactions. Several nucleases are thought to participate in the incision steps (second row), however, their exact sites of action are not defined. XPR/ERCC1, by analogy with NER, is thought to act 5′ of the ICLs, whereas the 3′ nuclease acting in RIR is not known (?). The FA pathway regulates the activity of nucleases in the replication-dependent pathway. FANCM facilitates fork regression whereas other FA proteins could recruit nucleases. Specific translesion synthesis nucleases participate in distinct repair pathways (third row): Polκ and Polζ in replication-independent repair and Rev1 and Polζ in the replication-dependent pathway. More than one DNA polymerase could be involved in the repair of an ICL. Replication-dependent ICL repair generates a DSB intermediate (bottom left), which is ultimately repaired via homologous recombination.