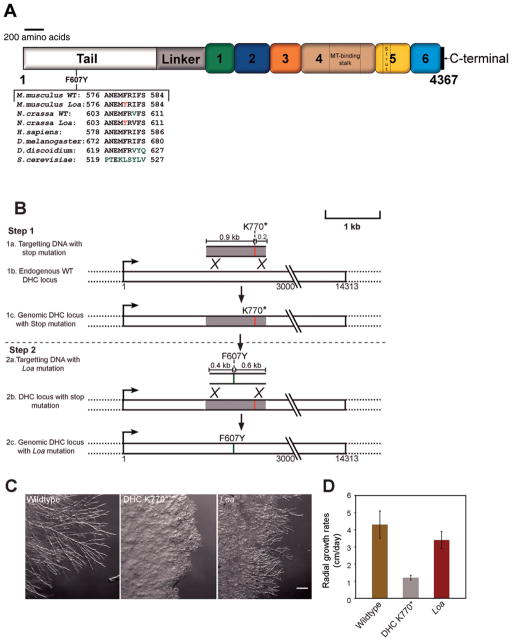
Fig. 1. Generation of an N. crassa Loa mutant strain.
A: Schematic representation of the cytoplasmic dynein heavy chain from N. crassa showing the position of the Loa mutation. Numbers 1–6 indicate appropriate AAA+ domains. MT and C represent the microtubule binding domain and C-terminal region respectively. The alignment of a subset of sequences shows the conservation of the DHC Loa mutation (F607Y) between N. crassa and other species. Black letters indicate conservation, green letters indicate altered amino acids, and red letters indicate the Loa mutation. B: A schematic representation of the two step strategy used to generate the Loa mutant strain. Step 1, introduction of a stop mutation to generate the ropy strain DHCK770*. Step 2, introduction of the Loa mutation and the rescue of the stop mutation. C: Comparison of colony morphologies between wildtype, mutant DHCK770* and Loa mutant strains. Note the Loa mutant strain exhibits mild ropy growth morphology in comparison to the wildtype strain. D: Bar graph showing radial growth rates of wildtype, mutant DHCK770* and Loa mutant strains. Data are shown as mean ± SD.