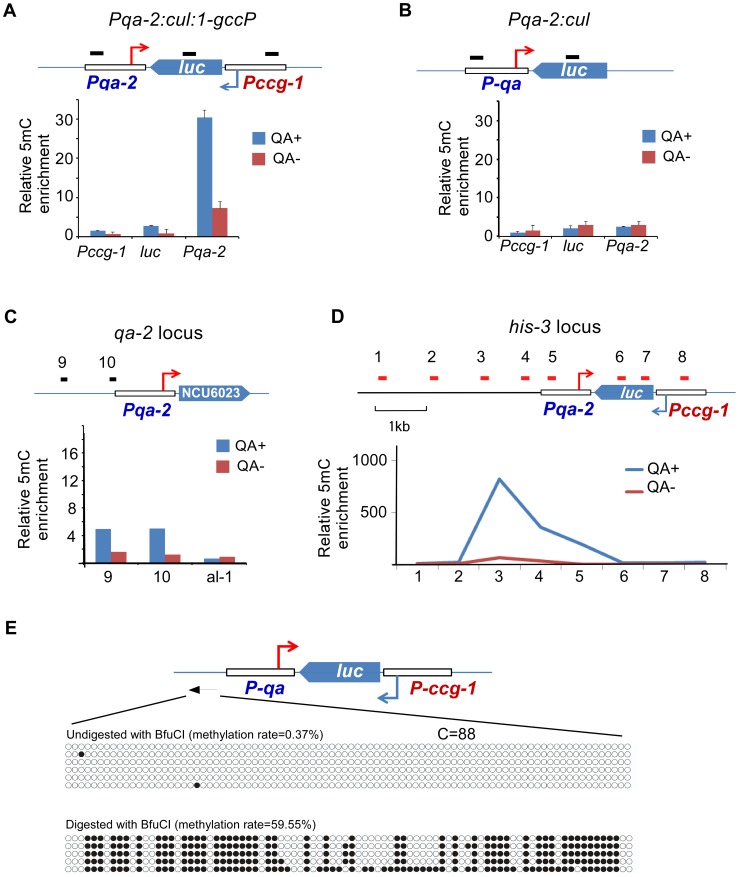
Figure 5. DNA methylation is induced in the promoter region of an artificial convergent transcription construct upon induction of transcription.
(A and B) MeDIP results showing the DNA methylation status of the ccg-1 promoter, luc gene body, and the qa-2 promoter of the indicated construct. The artificial construct Pqa-2:cul:1-gccP (A) or Pqa-2:cul (B) resides at the his-3 locus. The top cartoon of each panel depicts the architecture of the construct and black bars indicate the approximate location of primer sets. Three independent repeats were performed. Values are mean ± s.d. (C) Distribution of DNA methylation at the endogenous location of the qa-2 promoter of the Pqa-2:cul:1-gccP strain. (D) Distribution of DNA methylation around the Pqa-2:cul:1-gccP construct at the his-3 locus with/without the activation of the qa-2 promoter. In panels A-D, QA+ and QA- indicate presence or absence of quinic acid (QA), respectively. DNA methylation was measured with MeDIP. (E) Results of bisulfite PCR in the region upstream of the qa-2 promoter of the Pqa-2:cul:1-gccP construct. Two aliquots of genomic DNA from Pqa-2:cul:1-gccP strain, one digested with BfuCI and one untreated, were subject to bisulfite treatment and sequencing, respectively (strategy 2 of bisulfite sequencing, primer sequences in table S4). Each row of circles represents the order and number of cytosines in the subcloned sequence. Opened and filled circles indicate unmethylated and methylated cytosine, respectively.