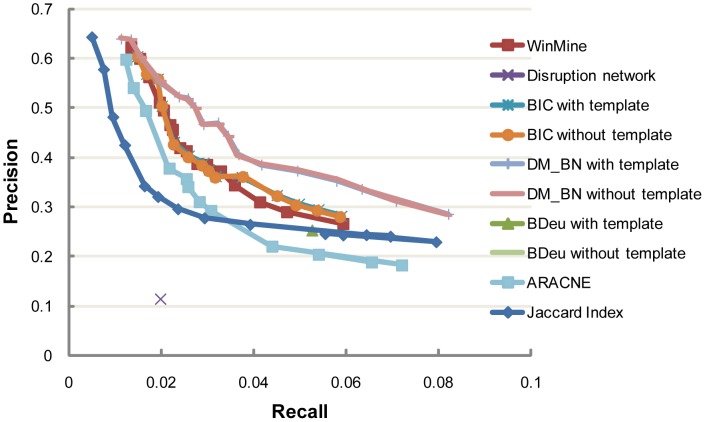
Figure 2. Precision and recall of networks inferred by DM_BN and by other network inference methods at various cutoffs.
The colored lines show the precision and recall tradeoffs as a function of different values of Jaccard index threshold, mutual information threshold in ARACNE, kappa parameter in WinMine, weight of the penalty term in BIC and the  parameter in DM_BN, respectively. The Disruption network approach and the BDeu metric do not have any tunable parameter and only one network can be predicted. We curated 5662 protein-protein interactions, regulatory interactions, genetic/epistatic interactions and protein complexes from the KEGG and SGD databases to evaluation the performance of different algorithms (See Table S2).
parameter in DM_BN, respectively. The Disruption network approach and the BDeu metric do not have any tunable parameter and only one network can be predicted. We curated 5662 protein-protein interactions, regulatory interactions, genetic/epistatic interactions and protein complexes from the KEGG and SGD databases to evaluation the performance of different algorithms (See Table S2).