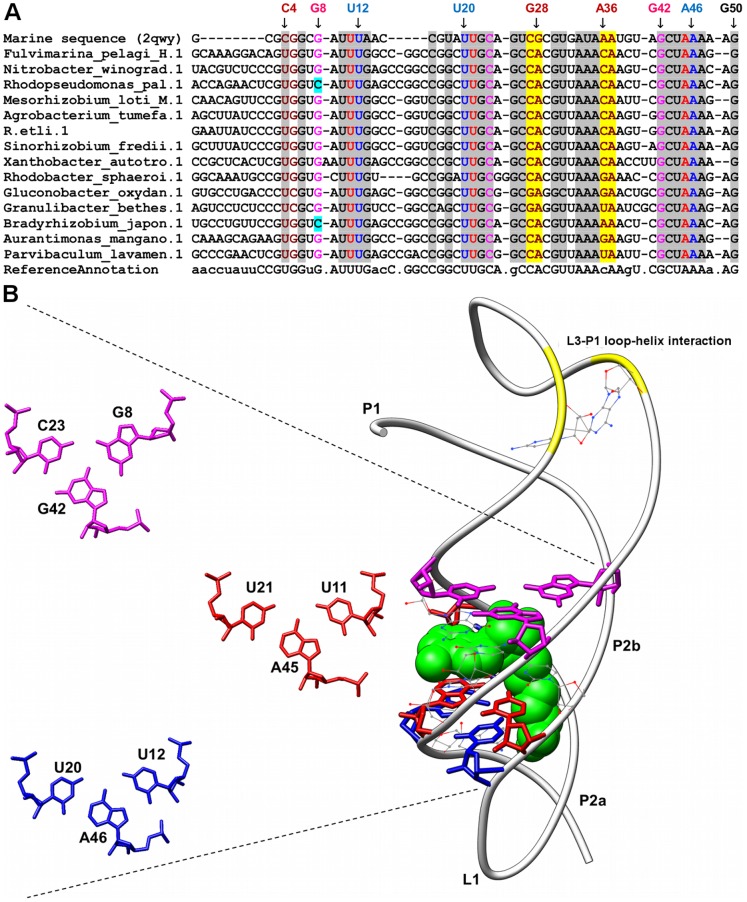
Figure 3. Sequence alignment and annotated tertiary motifs in SAM-II riboswitches.
(A) Sequence alignment of SAM-II riboswitches where gray shaded columns indicate the highly conserved nucleotide positions. Other color shaded columns indicate regions involved in tertiary interactions (brown: A-minor and yellow: ribose-zipper). For clarity, not all tertiary motif positions are numbered. Base-triple positions that are not conserved are in cyan shaded columns. (B) Tertiary structure of SAM-II riboswitch [13] (PDB ID: 2qwy). The ligand is represented as spheres and the highly conserved nucleotides are in wire representation. Magnification of the triples using numbering for 2qwy are presented in red (G8-C23-G42), magenta (U11-U21-A45), and blue (U12-U20-A46).