Figure 6. Sequence alignment and annotated tertiary motifs in lysine riboswitches.
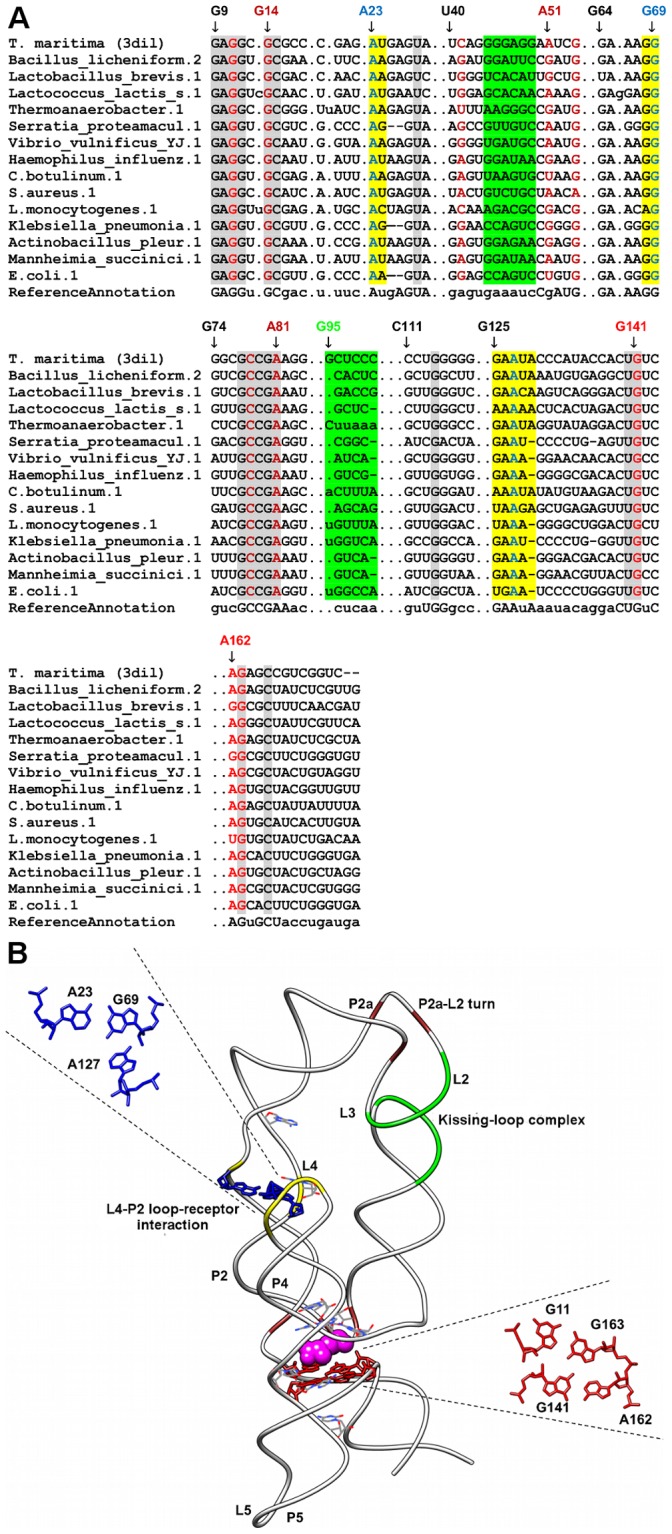
(A) Sequence alignment of lysine riboswitches where gray shaded columns indicate the highly conserved nucleotide positions. Other color shaded columns indicate regions involved in tertiary interactions (brown: A-minor, yellow: loop-receptor interaction and green: kissing-loop complex). For clarity, not all tertiary motif positions are numbered. Base-triple positions that are not conserved are in cyan shaded columns. (B) Tertiary structure of T. maritima lysine riboswitch [23] (PDB ID = 3dil). The ligand is represented as spheres and the highly conserved nucleotides are in wire representation. Base triple (A127-A23-G69) and base quadruple (G11-G163-G141-A162) interactions are presented in blue and red respectively.
