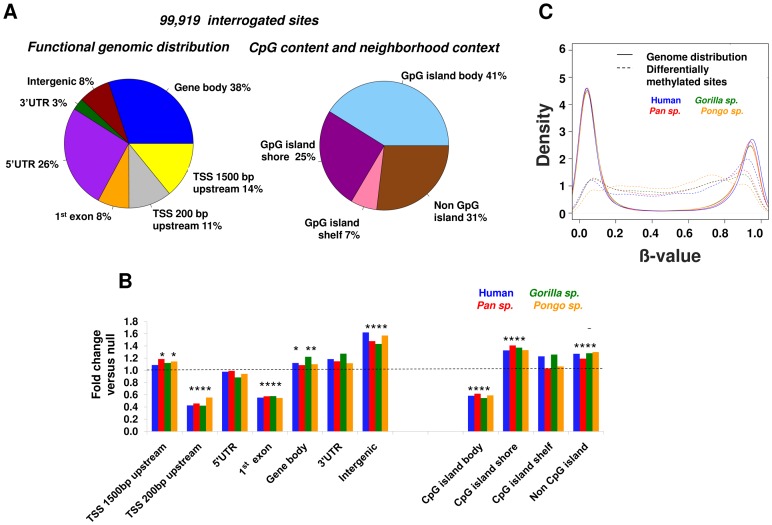
Figure 4. Location of differential methylation in primate genomes.
(A) Distribution of 99,191 CpG sites interrogated in all great ape species. Left: Gene-centric functional distribution of methylation changes : 1,500 bp upstream of gene TSSs, 200 bp upstream of TSSs, 5′UTR, 1st exon, gene body, 3′ UTR and intergenic. Right: CpG-island centric distribution: CpG island, shore (±2 kb flanking CpG islands), shelf (2–4 kb from CpG islands). (B) A non-random distribution of methylation changes in recent primate evolution. We observe an excess of differential CpG methylation within the first 1,500 bp upstream of gene TSSs, gene bodies, intergenic regions, shore regions flanking CpG islands and non-CpG island regions. In contrast DNA methylation tends to be relatively conserved close to gene transcription start sites (−200 bp of TSS to 1st exon), and in the body of CpG islands. Each bar shows the relative enrichment for differential methylation versus that expected under a null distribution. * denotes p<0.0001 (permutation test). (C) Density plot showing the distribution of methylation levels of differentially methylated sites compared to that in the rest of the genome. Sites of evolutionary change among great apes have a significantly different distribution (p = 2.2×10−16 Kolmogorov-Smirnov test).