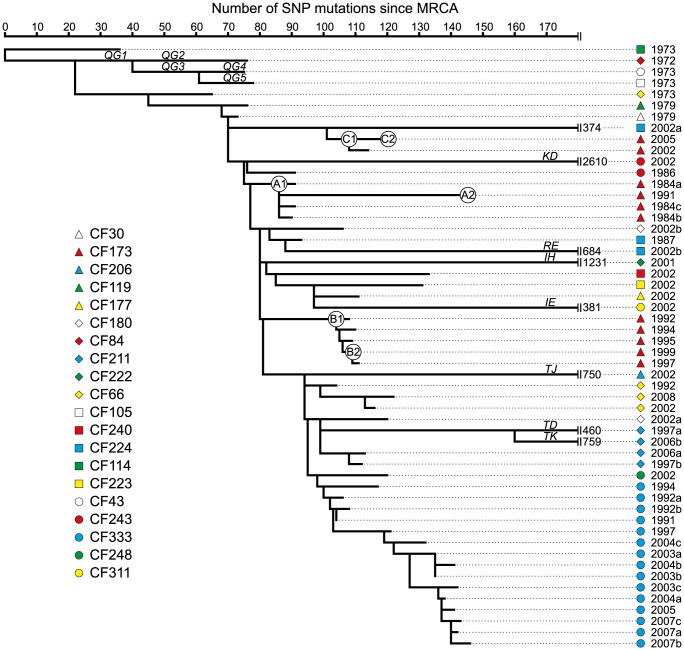
Figure 2. Maximum-parsimonious reconstruction of the phylogeny of the P. aeruginosa DK2 clones.
The phylogenetic tree is based on 7,326 SNPs identified from whole-genome sequencing, and lengths of branches are proportional to the number of mutations. Outlier isolate CF510-2006 [9] (not shown) was used as an outgroup to determine the root of the tree. Branches leading into mutS, mutL, or, mutY hypermutable isolates are named as indicated by italic letters. Statistics on mutations accumulated in the specific branches are summarized in Table 1. Circles labeled A1, A2, B1, B2, C1, and C2, respectively, denotes the position of the first and last genotype of each of the DK2 sub-lineages A, B, and C which were observed to have infected patient CF173.