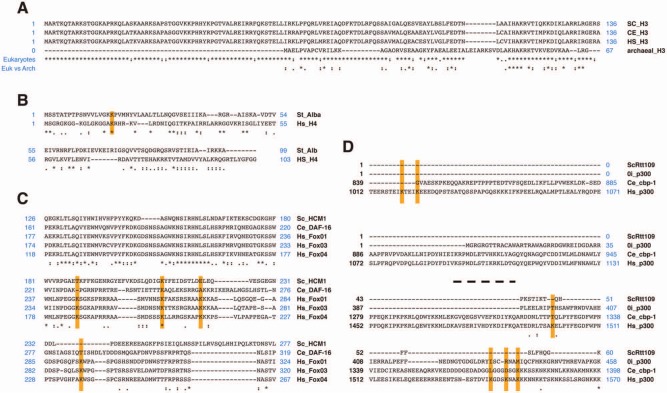
Figure 3.
Protein alignments using ClustalW. (A) Histone H3 alignment: alignment among eukaryotes versus archaea H3. (B) Alignment between archaea Alba protein and human histone H4. As it is shown, the sequence around the K16 residue is conserved in both cases. (C) FOXO protein alignment (fragment) from S. cerevisiae (Hcm1p) and C. elegans (Daf-16) to human homologs. The lysines known to be deacetylated by sirtuins in any of them are highlighted. (D) Alignment of p300 using S. cerevisiae (Rtt109p), Ostreococcus, C. elegans (cpb-1), and human homologs. Alignment of 2 different regions of the p300 homologs containing SIRT1 and SIRT2 lysine targets.