Abstract
Natural product biosynthesis has proven a fertile ground for the discovery of novel chemistry. Herein we review the progress made in elucidating the biosynthetic pathways of phosphonate and phosphinate natural products such as the antibacterial compounds dehydrophos and fosfomycin, the herbicidal phosphinothricin-containing peptides, and the antimalarial compound FR-900098. In each case, investigation of the pathway has yielded unusual, and often unprecedented, biochemistry. Likewise, recent investigations have uncovered novel ways to cleave the C-P bond to yield phosphate under phosphorus starvation conditions. These include the discovery of novel oxidative cleavage of the C-P bond catalyzed by PhnY and PhnZ as well as phosphonohydrolases that liberate phosphate from phosphonoacetate. Perhaps the crown jewel of phosphonate catabolism has been the recent resolution of the longstanding problem of the C-P lyase responsible for reductively cleaving the C-P bond of a number of different phosphonates to release phosphate. Taken together, the strides made on both metabolic and catabolic fronts illustrate an array of fascinating biochemistry.
Introduction
With resistance to commonly deployed antibiotics on the rise, new compounds are urgently needed to tackle pathogenic bacteria. Phosphonate and phosphinate natural products have been attractive as they are stable mimics of carboxylic acids and/or phosphate esters, thereby allowing them to potently inhibit enzymes in a variety of metabolic processes. Whereas phosphonates have only a single C-P bond, the less common phosphinates have either two C-P bonds or one C-P bond and a P-H bond [1,2]. The C-P bond can be broken in pathways dedicated to phosphonate catabolism as described in the second part of this review, but it is resistant to acid/base hydrolysis as well as the action of phosphatases and phosphodiesterases. Recent investigations of the pathways responsible for construction and breakdown of organophosphonates/phosphinates have yielded many novel enzymatic transformations [2–4]. This review will focus on pathways and enzymatic mechanisms that have been reported in the period 2009–2013, with particular emphasis on unusual chemistry.
Initial Steps in the Biosynthesis of Phosphonates: PEP Mutase and Phosphonopyruvate decarboxylase
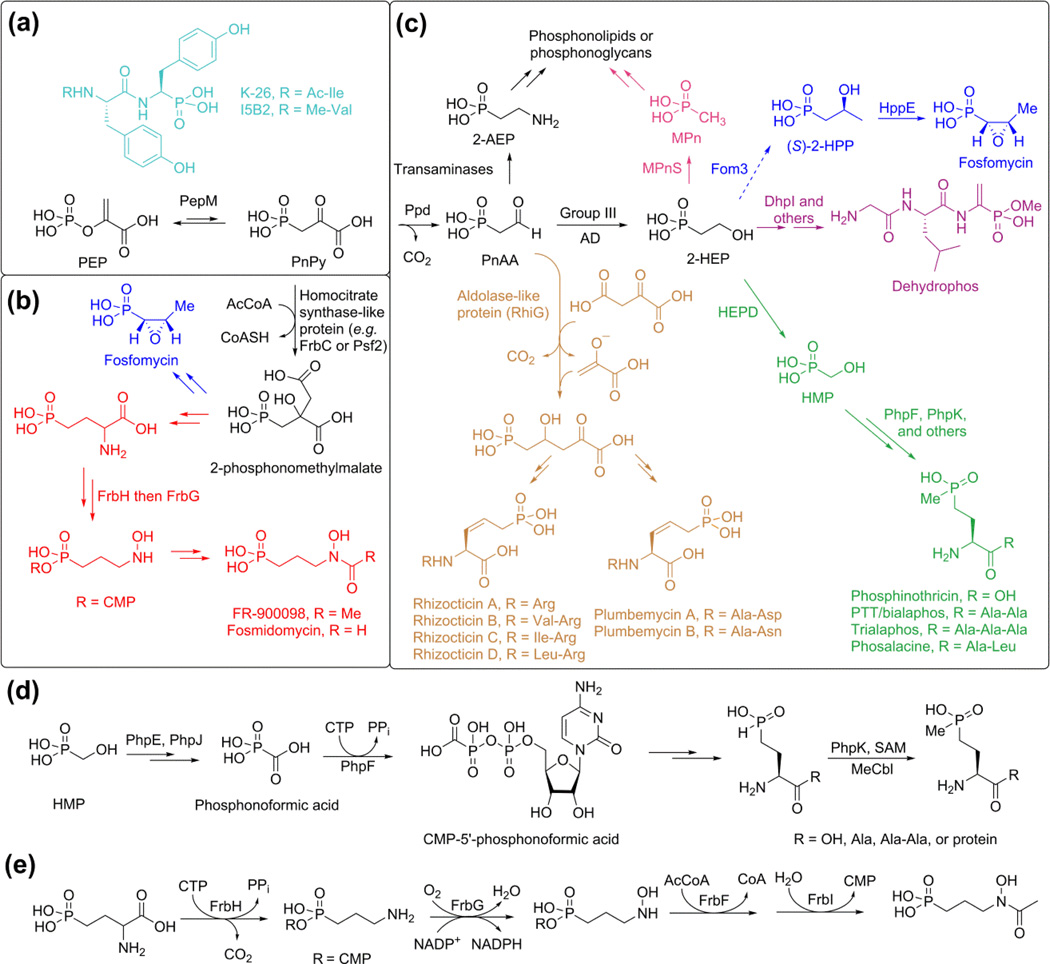
With the exception of the related phosphonotripeptides K-26 and I5B2 [5] (Figure 1a, light blue), the first step of all known phosphonate biosynthetic pathways is the reversible conversion of phosphoenolpyruvate (PEP) to phosphonopyruvate (PnPy) by PEP mutase (PepM) (Figure 1a). Because the isomerization equilibrium greatly favors PEP [6], PnPy formation is always connected to an essentially irreversible second step. One strategy is the addition of an acetate anion equivalent to the ketone of PnPy during the biosynthesis of FR-900098 and the fosfomycin biosynthetic pathway in pseudomonads (Figure 1b). A second, more common strategy involves decarboxylation of PnPy by phosphonopyruvate decarboxylase (ppd) to produce phosphonoacetaldehyde (PnAA), which is subsequently functionalized in myriad ways (Figure 1c). These transformations include reduction by metal dependent alcohol dehydrogenases (ADs) to generate 2-hydroxyethyl phosphonate (2-HEP) [7] and transamination to produce the polar head group for phosphonoglycans or phosphonolipids [8] (Figure 1c).
Figure 1.
Overview of phosphonate biosynthesis. Frequently encountered reactions are shown in black in panels a-c. Unique transformations towards various family members are shown in color. Because of space constraints, the rhizocticin/plumbemycin pathway (light brown) [9] will not be discussed in this review. (a) All known pathways start with the conversion of PEP to PnPy except for the biosynthesis of K-26 and I5B2. (b) During the biosynthesis of FR-900098 in actinomycetes and fosfomycin in pseudomonads, an acetyl group is added to the ketone of PnPy. (c) A range of products are accessed from PnAA after decarboxylation of PnPy. (d) Biochemically intriguing steps in phosphinothricin biosynthesis. The physiological substrate for the P-methylase is currently unknown. (e) The late stages of FR-900098 biosynthesis.
Dehydrophos
The antibiotic dehydrophos is currently the only known naturally occurring phosphonate that is esterified; it also contains an unusual aminovinylphosphonate moiety [10] (Figure 1b, purple). Both functionalities are critical for its antimicrobial activity [11]. Like many phosphonopeptides, dehydrophos is a Trojan horse antibiotic that is taken up by peptide permeases and activated by intracellular proteolysis, which unmasks methyl acetylphosphonate that potently inhibits pyruvate dehydrogenase [11,12] and 1-deoxy-D-xylulose-5-phosphate synthase [13]. Production of dehydrophos in a heterologous host identified sixteen genes that were required for dehydrophos production [12]. Comparison to homologous proteins of known function, construction of genetic knockouts, and in vitro characterization of nine proteins allowed construction of a hypothetical pathway that involves the use of aminoacylated tRNA for non-ribosomal peptide bond formation [7,12,14,15]. The S-adenosyl methionine (SAM) dependent protein responsible for O-methylation of dehydrophos, DhpI, is intriguing for both its unusual "domain-swapped dimer" architecture as well as its relaxed substrate specificity [14].
Phosphinothricin
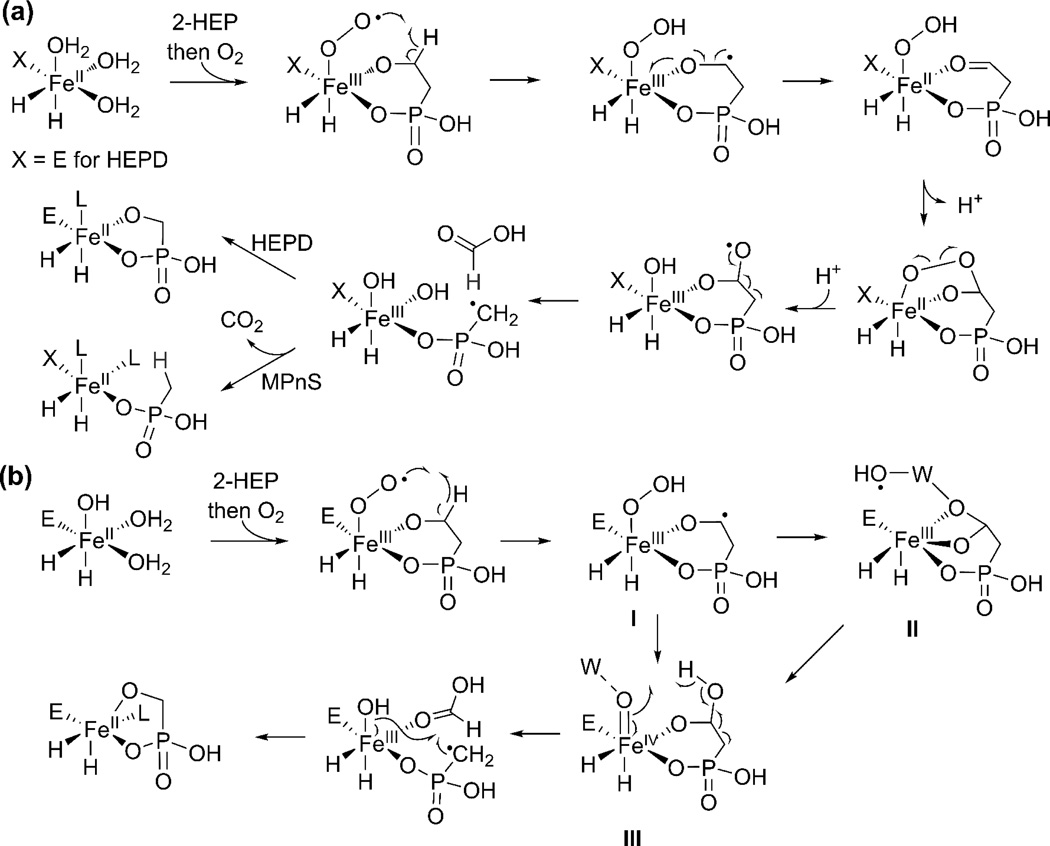
Another class of Trojan horse compounds is the phosphinothricin (PT) containing peptides (Figure 1c, green) such as phosphinothricin tripeptide (PTT). PT is the active compound in a number of commercially important herbicides such as Liberty and Basta. Identification of the gene cluster responsible for PTT production enabled the in vitro characterization of a number of unusual reactions [16–18]. These include the scission of the unactivated carbon-carbon bond in 2-HEP to afford hydroxymethylphosphonate (HMP) and formate catalyzed by 2-HEP dioxygenase (HEPD) (Figure 1c), as well as the ligation of CMP to phosphonoformate by PhpF (Figure 1d) [17,19]. Mechanistic investigations of the non-heme iron dependent enzyme HEPD with isotopically labeled substrates have demonstrated that the enzyme abstracts the pro-S hydrogen from C2, and that the reaction takes place with loss of stereochemistry at C1 (Figure 2a) [20]. Because the enzyme does not require any external electron source, a ferric superoxo species is believed to abstract the hydrogen atom from C2 [21], a step that may be facilitated by the bidentate binding of 2-HEP observed in a cocrystal structure [19]. Density functional theory calculations have suggested a mechanism in which a bridged peroxide is cleaved homolytically (Figure 2a), which after β-scission results in a radical intermediate that explains the loss of stereochemistry at C1 [21]. Alternatively a mechanism involving an Fe(IV)-oxo intermediate was proposed that also results in the same radical (Figure 2b) [22]. The Fe(IV)-oxo was postulated to be formed via one of several possible mechanisms. One of these pathways involves a hydroxyl radical, but such an intermediate is typically of very high energy and appears less likely.
Figure 2.
Proposed mechanisms of catalysis by the structurally-related enzymes HEPD and MPnS. (a) HEPD and MPnS are hypothesized to share a consensus mechanism that diverges only in the final step, with a radical rebound type hydroxylation for HEPD and hydrogen atom abstraction from formate for MPnS. (b) DFT calculations suggest that HEPD could proceed via an alternative mechanism that either indirectly (hydroperoxo intermediate I to radical II, and then to ferryl III) or directly (I to III) leads to a ferryl intermediate that abstracts a hydrogen atom from a hydroxyl group. A similar alternative mechanism with hydrogen atom abstraction from formate as the last step can also be drawn for MPnS. W is a water molecule; L is an unknown ligand.
Recently, the intriguing P-methylase (PhpK) from the phosalacine pathway (another PT containing natural product, Figure 1c) was successfully reconstituted and demonstrated to belong to the class of radical-SAM enzymes that contain a [Fe-S] cluster but also require methylcobalamin (Figure 1d) [23]. While the mechanism of PhpK is still unresolved, the success in reconstituting this enzymatic transformation suggests it might be possible to activate similar enzymes, such as the putative methyltransferase Fom3 involved in fosfomycin biosynthesis (see below).
Fosfomycin
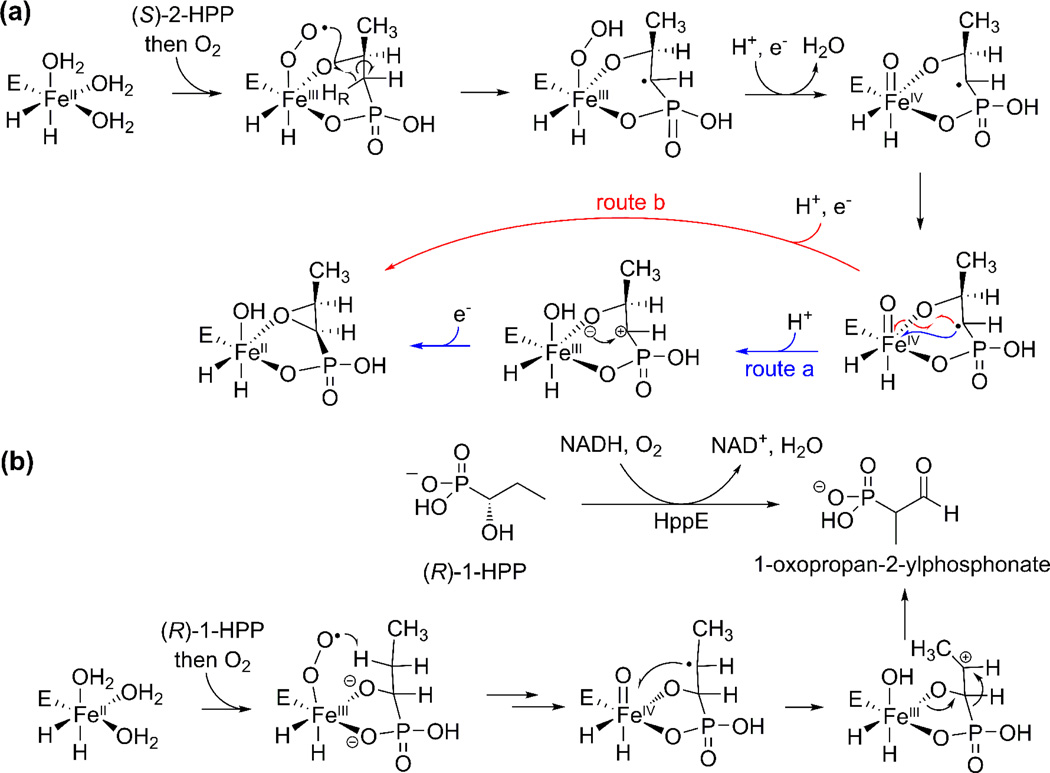
Fosfomycin, made by both streptomycetes and pseudomonads (Figure 1b and 1c, blue), inhibits cell-wall biosynthesis and is used clinically to treat infections. Based on the gene cluster and feeding experiments in streptomyces [24], it has been proposed that a methyl group is inserted into a C-H bond at C2 of 2-HEP to form (S)-2-hydroxpropylphosphonate (2-HPP) by the protein encoded by fom3 (Figure 1c) [25,26]. Fom3 has homology with PhpK discussed above, but at present its activity has yet to be reconstituted in vitro. Because the unreactive C-H bond of 2-HEP is not nucleophilic and because the Fom3 sequence suggests it is a member of the radical-SAM family, it is anticipated that this reaction will proceed via novel radical-mediated chemistry [25,26]. The final step in fosfomycin biosynthesis is the epoxidation carried out by 2-HPP epoxidase (HppE) [27]. This enzyme is unusual because epoxidations usually take place with addition of an oxygen atom of O2 to a double bond, but HppE catalyzes a net dehydrogenation and inserts the alcohol oxygen from 2-HPP [28] (Figure 3a). Two of the electrons required to reduce O2 come from 2-HPP with the other two electrons coming from a reductase. Interestingly, HppE has very little sequence similarity to HEPD from PTT biosynthesis, but quite striking structural homology [19,29]. The recent demonstration that HppE oxidizes the substrate analog (R)-1-HPP to 1-oxopropan-2-ylphosphonate (Figure 3b), likely via a carbocation intermediate, raises the question whether the oxidation of (S)-2-HPP might also proceed through a similar carbocation intermediate [30](Figure 3a, route a).
Figure 3.
Proposed mechanisms for reactions catalyzed by HppE. (a) Transformation of (S)-2-HPP to fosfomycin either involves a carbocation intermediate (route a) or takes place by an intramolecular oxygen atom rebound-like mechanism (route b). (b) Evidence for the carbocation route in panel (a) is based on the observation that HppE converts (R)-1-HPP to 1-oxopropan-2-ylphosphonate, likely via a carbocation intermediate.
Recently, a very different pathway for fosfomycin biosynthesis was reported in Pseudomonas syringae [31]. Although the biosynthetic gene cluster harbors genes encoding both a PEP mutase and an HppE homolog, no other proteins were homologous to those encoded by the fosfomycin gene clusters of several streptomycetes. For example, no ppd was present and instead, a citrate synthase-like protein likely catalyzes the addition of an acetate anion equivalent to PnPy to generate 2-phosphonomethylmalate [31] (Figure 1b), similar to the pathway to FR-900098 (see below).
FR-900098 and Fosmidomycin
FR-900098 and fosmidomycin (Figure 1b, red) are related phosphonates with potent antimalarial activities due to inhibition of 1-deoxy-D-xylulose 5-phosphate reductoisomerase in the nonmevalonate pathway of isoprenoid biosynthesis. The FR-900098 biosynthetic enzymes FrbD (PEP mutase) and FrbC in vitro converted PEP to 2-phosphonomethylmalate in the presence of acetyl-CoA (Figure 1b) [32], the first time a modification other than decarboxylation had been observed as the driving force for PnPy formation.
The late steps of FR-900098 biosynthesis have also been characterized in vitro [33,34]. Perhaps the most interesting enzyme is the bifunctional enzyme FrbH. The nucleotide transferase domain is responsible for ligating CMP to the phosphonate group (as was observed in PTT biosynthesis, Figure 1d), and the PLP-binding domain then facilitates decarboxylation (Figure 1e) [33]. Feeding experiments and in vitro experiments established that FrbG subsequently catalyzes the flavin-dependent N-hydroxylation followed by N-acetylation by FrbF in the presence of acetyl-CoA [33] to ultimately furnish FR-900098 (Figure 1b, red).
Methylphosphonate Synthesis: A Link to C-P Lyases
Methane and oxygen concentrations are both supersaturating in the surface waters of the oceans, which is surprising since biological methane formation is an anaerobic activity. A recently discovered phosphonate pathway in the archeon Nitrosopumilus maritimus might provide an explanation for this oceanic methane paradox [35]. It had been suggested that methane might be formed under aerobic conditions from methylphosphonate by the action of C-P lyase (see below) [36], but methylphosphonate was not a known naturally occurring compound. Genome mining for phosphonate biosynthetic gene clusters showed N. maritimus harbored the canonical pathway for formation of 2-HEP (Figure 1a, magenta) [35]. One other gene in the cluster encoded a protein with homology to HEPD (discussed above). When the protein was expressed, reconstituted with Fe(II), and incubated with 2-HEP, the observed product was not HMP (the product of HEPD) but rather methylphosphonate (MPn), and the enzyme was named methylphosphonate synthase (MPnS). NMR analysis of cell extracts of N. maritimus indicated that the methylphosphonate is likely esterified to a larger biomolecule, similar to how 2-AEP decorates biopolymers in bacteria and lower eukaryotes [8,35].
Like HppE and HEPD, MPnS uses novel oxidative chemistry, in this case to cleave the carbon-carbon bond of 2-HEP to generate methylphosphonate (MPn) and CO2 (Figure 2a) [35]. Despite the low sequence identity between HEPD and MPnS (21%), critical residues such as those responsible for Fe(II) and substrate binding are conserved [37]. Substrate analog studies have indicated that MPnS and HEPD both abstract the pro-S C2 hydrogen from 2-HEP to initiate catalysis [20,37]. Furthermore, the incipient proton on the methyl group of MPn derives not from solvent, but from the pro-R hydrogen at C2 of 2-HEP [37]. This observation has led to the proposal that MPnS and HEPD share a consensus mechanism that differs only in the final step of catalysis, which involves a hydroxyl rebound-like step for HEPD, and abstraction of a hydrogen atom from formate for MPnS (Figure 2a; a similar branched mechanism can also be drawn for the mechanism in Figure 2b).
Radically Different: C-P Lyase Catabolism
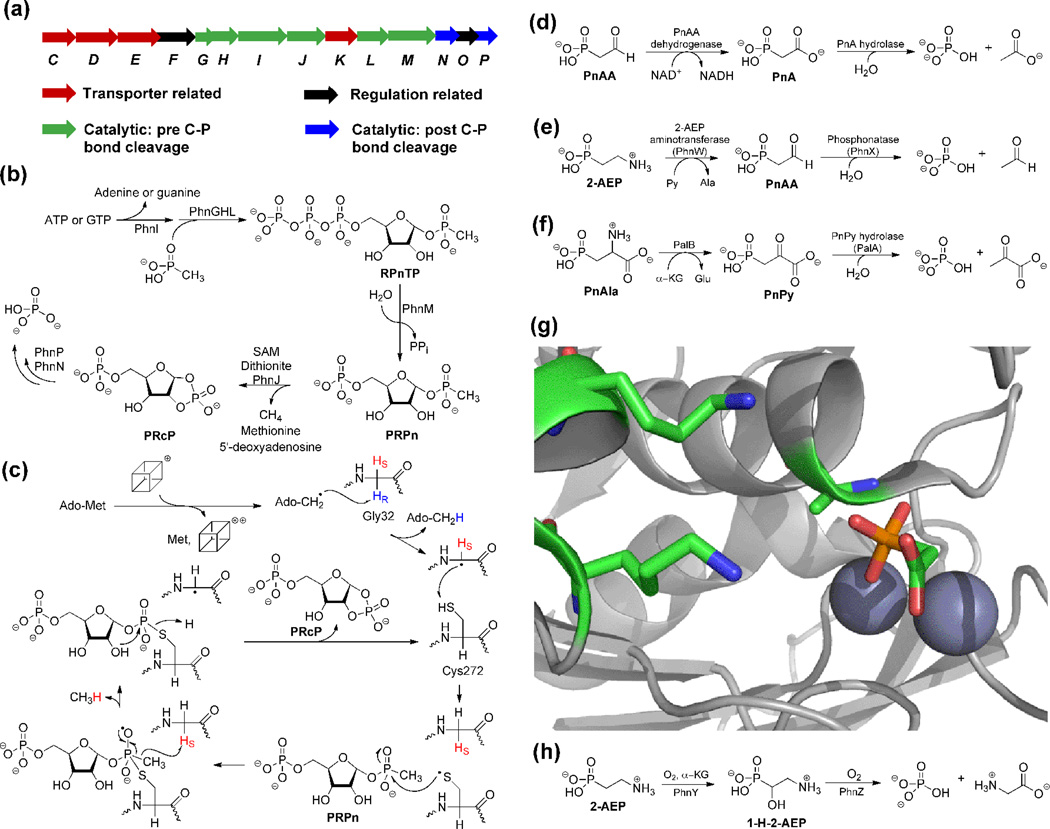
Phosphorus is a limiting nutrient in many environments, and hence organisms have evolved pathways that can liberate phosphate from various naturally occurring phosphonates. Several strategies have been uncovered including hydrolytic, oxidative, and reductive processes (Figure 4). The last of these is catalyzed by the until recently enigmatic C-P lyase. Ever since the genes responsible for C-P lyase activity were discovered in E. coli (Figure 4a) [39], numerous efforts had been made to reconstitute the activity of the system in vitro. However, until recently, characterization of proteins in the cluster had been confined to either regulatory proteins such as PhnO [40] or proteins catalyzing reactions after C-P bond cleavage [41–44]. Zechel, Hove-Jensen, and coworkers were able to demonstrate via copurification and crosslinking assays that PhnGHIJK very likely formed a multiprotein complex [45], but they were unable to demonstrate C-P lyase activity in vitro.
Figure 4.
Overview of phosphonate catabolism. (a) Gene cluster of the Phn operon for reductive C-P bond cleavage. (b) Schematic overview of the C-P lyase pathway. (c) A recently proposed mechanism for PhnJ catalyzed C-P bond cleavage [38]. The cube represents the [4Fe-4S] cluster. (d) Phosphonoacetate hydrolase (PhnA) is a newly discovered hydrolase that takes advantage of the carboxylate group as a "β electron sink". (e) Phosphonatase is able to convert 2-AEP to free phosphate through PnAA. (f) PnPy hydrolase is used to liberate phosphate from PnPy after transamination of PnAla. (g) The two lysines (shown as sticks) in the PhnA-T68A active site are relatively far from PnA. Ala68 is set up for in-line attack on the phosphonate when this residue is Thr in wild type PhnA, and the π-system of the carboxylate of PnA is well oriented to accept electron density upon cleavage of the P-C bond. The incipient negative charge may also be stabilized by binding to a Zn(II) in a monodentate fashion. (h) A recently discovered pathway from a genomic marine library results in oxidative cleavage of the C-P bond.
More recently, the initial steps in the pathway were reconstituted in vitro by Raushel and coworkers. They showed that ligation of methylphosphonate to ribose triphosphate (RTP) produces ribose 1-methylphosphonate 5-triphosphate (RPnTP) when PhnGHL were all expressed as fusion proteins to ensure solubility and after these solubility tags were removed in situ [46]. PhnM then converted RPnTP to ribose 1-methylphosphonate 5-phosphate (PRPn) [46], which had been previously proposed as the substrate for the C-P lyase PhnJ [45] (Figure 4b).
PhnJ is a radical SAM enzyme that converts SAM and PRPn to PRcP, methane and the SAM-cleavage products methionine and 5'-deoxyadenosine [46]. These observations as well as subsequent mechanistic studies have led to a proposed mechanism for the fascinating reaction catalyzed by PhnJ (Figure 4c; for more details see the article by Raushel and coworkers in this issue[v1]) [38,46,47]. The production of PRcP by PhnJ would allow PhnP and downstream proteins to liberate phosphorus as inorganic phosphate for primary metabolic purposes [41,43,44].
Non CP-Lyase Catabolism: β Electron Sinks
In addition to the C-P lyase strategy, another approach to liberate phosphorus makes use of phosphonates with a "β electron sink" (Figure 4d–f). In these substrates, the C-P bond is broken hydrolytically, which is facilitated by either a carbonyl (or Schiff base equivalent) or carboxylate moiety that is able to stabilize incipient negative charge on the α-carbon. The active sites of the hydrolytic enzymes also aid in stabilization of the delocalized negative charge through hydrogen bonding or use of a positively charged metal ion. PnAA hydrolase, also known as phosphonatase, and PnPy hydrolase have been well-studied [48–51] (Figure 4e and 4f) and will not be discussed in detail here. Instead, we will focus on another phosphonohydrolase that has more recently been discovered, phosphonoacetate hydrolase (PhnA).
In organisms that lack PnAA and PnPy hydrolase, 2-AEP can be transaminated to PnAA, which can be oxidized to PnA by phosphonoacetaldehyde dehydrogenase [52,53] (Figure 4d). PnA hydrolase is a member of the alkaline phosphatase (AP) family and like other members of the family, the active site contains two divalent metal ions and a catalytic Thr that is believed to carry out in-line attack on the phosphonate [54,55]. However, the leaving group for this reaction (an enolate) is quite different than those of the typical alkaline phosphatase family members (oxygen based anions). How PnA hydrolase stabilizes the incipient delocalized negative charge as a result of C-P bond scission is still incompletely understood. It was initially proposed that two conserved Lys residues might stabilize the enediolate dianion through hydrogen bonding and electrostatic interactions [54]. More recently, crystal structures of a PnA hydrolase variant in which the Thr was mutated to Ala with PnA bound demonstrated monodentate binding of the acetate group of the substrate to one of the two metal ions, with the C-P bond to be broken and the π system of the acetate group well oriented for charge delocalization (Figure 4g) [55]. This result led to a proposal that the divalent metal ion helps stabilize the dianion in a manner reminiscent of mandelate racemase [56].
Non CP-Lyase Catabolism: Oxidative Cleavage
Screening of a genomic marine library for fosmids that allowed a C-P lyase deficient strain of E. coli to grow on phosphonates led to the discovery of two new enzymes capable of phosphonate catabolism, PhnY and PhnZ (Figure 4h) [57]. Neither enzyme has homology to any enzyme known to be involved in phosphonate metabolism. PhnY is an Fe(II)/α-ketoglutarate dependent enzyme that stereospecifically hydroxylates 2-AEP at the C1 position to afford 1-hydroxy-2-aminoethylphosphonate (1-H-2-AEP) [58]. PhnZ subsequently uses O2 to oxidize 1-H-2-AEP to phosphate and glycine [58]. PhnZ was proposed to be a dinuclear Fe-dependent enzyme [58] that might affect catalysis in a similar manner as myo-inositol oxygenase [59].
Summary and Outlook
Investigations of the biochemistry of phosphonate biosynthesis over the past five years have uncovered a myriad of novel enzymatic transformations. Likewise, the discovery of additional catabolic pathways and the strides made in reconstituting the activity of previously intractable enzymes have provided access to intriguing and unusual enzymology. Hence, both anabolism and catabolism of phosphonates have turned out to be a treasure trove of unusual chemistry.
Phosphonates inhibit enzymes by mimicking phosphate esters or carboxylates present in their substrates
Phosphonate biosynthetic and catabolic pathways contain many unusual chemical transformations
Organisms have evolved at least three different strategies to utilize phosphonates as phosphate source
Discovery of methylphosphonate synthase provides an explanation for supersaturated methane in the aerobic oceans
C-P lyase is an unusual radical-SAM protein that cleaves the C-P bond of unactivated phosphonates such as methylphosphonate
Acknowledgment
The authors acknowledge Dr. Despina Bougioukou (UIUC) for critically reviewing the manuscript and Dr. Qi Zhang (UIUC) for help assembling Figure 4. This work was supported by the National Institute of Health (GM PO1GM077596).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Recommended Reading
Papers of Outstanding Interest
Papers of Special Interest
- 1.White AK, Metcalf WW. Microbial metabolism of reduced phosphorus compounds. Annu Rev Microbiol. 2007;61:379–400. doi: 10.1146/annurev.micro.61.080706.093357. [DOI] [PubMed] [Google Scholar]
- 2.Metcalf WW, van der Donk WA. Biosynthesis of Phosphonic and Phosphinic Acid Natural Products. Annu Rev Biochem. 2009;78:65–94. doi: 10.1146/annurev.biochem.78.091707.100215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nair SK, van der Donk WA. Structure and mechanism of enzymes involved in biosynthesis and breakdown of the phosphonates fosfomycin, dehydrophos, and phosphinothricin. Arch Biochem Biophys. 2011;505:13–21. doi: 10.1016/j.abb.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Villarreal-Chiu JF, Quinn JP, McGrath JW. The genes and enzymes of phosphonate metabolism by bacteria, and their distribution in the marine environment. Frontiers in Microbiology. 2012;3:1. doi: 10.3389/fmicb.2012.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ntai I, Manier ML, Hachey DL, Bachmann BO. Biosynthetic origins of C-P bond containing tripeptide K-26. Org Lett. 2005;7:2763–2765. doi: 10.1021/ol051091d. Compelling evidence is provided via isotope labeling that the K-26 phosphonate tripeptide is biosynthesized via a pathway that does not proceed through the canonical pathway that starts with PEP mutase. The alternative biosynthetic route to K-26 is still unresolved.
- 6.Bowman E, McQueney M, Barry RJ, Dunaway-Mariano D. Catalysis and Thermodynamics of the Phosphoenolpyruvate/Phosphopyruvate Rearrangement. Entry into the Phosphonate Class of Naturally Occurring Organophosphorus Compounds. J Am Chem Soc. 1988;110:5575–5576. [Google Scholar]
- 7.Shao Z, Blodgett JA, Circello BT, Eliot AC, Woodyer R, Li G, van der Donk WA, Metcalf WW, Zhao H. Biosynthesis of 2-hydroxyethylphosphonate, an unexpected intermediate common to multiple phosphonate biosynthetic pathways. J Biol Chem. 2008;283:23161–23168. doi: 10.1074/jbc.M801788200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hilderbrand RL. The Role of Phosphonates in Living Systems. Boca Raton: CRC Press; 1983. [Google Scholar]
- 9.Borisova SA, Circello BT, Zhang JK, van der Donk WA, Metcalf WW. Biosynthesis of rhizocticins, antifungal phosphonate oligopeptides produced by Bacillus subtilis ATCC6633. Chem Biol. 2010;17:28–37. doi: 10.1016/j.chembiol.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whitteck JT, Ni W, Griffin BM, Eliot AC, Thomas PM, Kelleher NL, Metcalf WW, van der Donk WA. Reassignment of the structure of the antibiotic A53868 reveals an unusual amino dehydrophosphonic acid. Angew Chem Int Ed. 2007;46:9089–9092. doi: 10.1002/anie.200703810. [DOI] [PubMed] [Google Scholar]
- 11.Kuemin M, van der Donk WA. Structure-activity relationships of the phosphonate antibiotic dehydrophos. Chem Commun. 2010;46:7694–7696. doi: 10.1039/c0cc02958k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Circello BT, Eliot AC, Lee JH, van der Donk WA, Metcalf WW. Molecular cloning and heterologous expression of the dehydrophos biosynthetic gene cluster. Chem Biol. 2010;17:402–411. doi: 10.1016/j.chembiol.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brammer LA, Smith JM, Wade H, Meyers CF. 1-Deoxy-d-xylulose 5-Phosphate Synthase Catalyzes a Novel Random Sequential Mechanism. J Biol Chem. 2011;286:36522–36531. doi: 10.1074/jbc.M111.259747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee JH, Bae B, Kuemin M, Circello BT, Metcalf WW, Nair SK, van der Donk WA. Characterization and structure of DhpI, a phosphonate O-methyltransferase involved in dehydrophos biosynthesis. Proc Natl Acad Sci U S A. 2010;107:17557–17562. doi: 10.1073/pnas.1006848107. This paper reports the unusual architecture and activity of the O-methyltransferase (DhpI) involved in dehydrophos biosynthesis.
- 15.Bougioukou DJ, Mukherjee S, van der Donk WA. Revisiting the biosynthesis of dehydrophos reveals a tRNA-dependent pathway. Proc Natl Acad Sci USA. 2013 doi: 10.1073/pnas.1303568110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blodgett JA, Zhang JK, Metcalf WW. Molecular cloning, sequence analysis, and heterologous expression of the phosphinothricin tripeptide biosynthetic gene cluster from Streptomyces viridochromogenes DSM 40736. Antimicrob Agents Chemother. 2005;49:230–240. doi: 10.1128/AAC.49.1.230-240.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blodgett JA, Thomas PM, Li G, Velasquez JE, van der Donk WA, Kelleher NL, Metcalf WW. Unusual transformations in the biosynthesis of the antibiotic phosphinothricin tripeptide. Nat Chem Biol. 2007;3:480–485. doi: 10.1038/nchembio.2007.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwartz D, Berger S, Heinzelmann E, Muschko K, Welzel K, Wohlleben W. Biosynthetic gene cluster of the herbicide phosphinothricin tripeptide from Streptomyces viridochromogenes Tu494. Appl Environ Microbiol. 2004;70:7093–7102. doi: 10.1128/AEM.70.12.7093-7102.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cicchillo RM, Zhang H, Blodgett JAV, Whitteck JT, Li G, Nair SK, van der Donk WA, Metcalf WW. An unusual carbon-carbon bond cleavage reaction during phosphinothricin biosynthesis. Nature. 2009;459:871–874. doi: 10.1038/nature07972. This study reports the crystal structure of HEPD as well as initial investigations into the mechanism of the unusual reaction it catalyzes.
- 20.Whitteck JT, Malova P, Peck SC, Cicchillo RM, Hammerschmidt F, van der Donk WA. On the stereochemistry of 2-hydroxyethylphosphonate dioxygenase. J Am Chem Soc. 2011;133:4236–4239. doi: 10.1021/ja1113326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirao H, Morokuma K. Ferric Superoxide and Ferric Hydroxide Are Used in the Catalytic Mechanism of Hydroxyethylphosphonate Dioxygenase: A Density Functional Theory Investigation. J Am Chem Soc. 2010;132:17901–17909. doi: 10.1021/ja108174d. [DOI] [PubMed] [Google Scholar]
- 22.Du L, Gao J, Liu Y, Liu C. Water-Dependent Reaction Pathways: An Essential Factor for the Catalysis in HEPD Enzyme. J Phys Chem B. 2012;116:11837–11844. doi: 10.1021/jp305454m. [DOI] [PubMed] [Google Scholar]
- 23. Werner WJ, Allen KD, Hu K, Helms GL, Chen BS, Wang SC. In Vitro Phosphinate Methylation by PhpK from Kitasatospora phosalacinea. Biochemistry. 2011;50:8986–8988. doi: 10.1021/bi201220r. This study on the P-methyl transferase involved in phosphinothricin biosynthesis reports the first successful in vitro reconstitution of a vitamin B12-dependent radical-SAM methyltransferase.
- 24.Seto H, Kuzuyama T. Bioactive Natural Products with Carbon-Phosphorus Bonds and Their Biosynthesis. Nat Prod Rep. 1999;16:589–596. doi: 10.1039/a809398i. [DOI] [PubMed] [Google Scholar]
- 25.Woodyer RD, Li G, Zhao H, van der Donk WA. New insight into the mechanism of methyl transfer during the biosynthesis of fosfomycin. Chem Commun. 2007:359–361. doi: 10.1039/b614678c. [DOI] [PubMed] [Google Scholar]
- 26.Booker SJ. Anaerobic functionalization of unactivated C-H bonds. Curr Opin Chem Biol. 2009;13:58–73. doi: 10.1016/j.cbpa.2009.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu P, Murakami K, Seki T, He X, Yeung SM, Kuzuyama T, Seto H, Liu HW. Protein purification and function assignment of the epoxidase catalyzing the formation of fosfomycin. J Am Chem Soc. 2001;123:4619–4620. doi: 10.1021/ja004153y. [DOI] [PubMed] [Google Scholar]
- 28.Hammerschmidt F. Biosynthesis of Natural Products with a P-C Bond. Part 8: On the Origin of the Oxirane Oxygen Atom of Fosfomycin in Streptomyces fradiae. J Chem Soc Perkin Trans 1. 1991:1993–1996. [Google Scholar]
- 29.Higgins LJ, Yan F, Liu P, Liu HW, Drennan CL. Structural insight into antibiotic fosfomycin biosynthesis by a mononuclear iron enzyme. Nature. 2005;437:838–844. doi: 10.1038/nature03924. [DOI] [PubMed] [Google Scholar]
- 30. Chang Wc, Dey M, Liu P, Mansoorabadi SO, Moon SJ, Zhao ZK, Drennan CL, Liu Hw. Mechanistic studies of an unprecedented enzyme-catalysed 1,2-phosphono-migration reaction. Nature. 2013;496:114–118. doi: 10.1038/nature11998. This report details the results of incubation of an unnatural substrate analog with HppE. Surprisingly, the transformation likely proceeds through a carbocation intermediate, raising questions about whether this might be the case for oxidation of the natural substrate of HppE.
- 31. Kim SY, Ju KS, Metcalf WW, Evans BS, Kuzuyama T, van der Donk WA. Different Biosynthetic Pathways to Fosfomycin in Pseudomonas syringae and Streptomyces Species. Antimicrob Agents Chemother. 2012;56:4175–4183. doi: 10.1128/AAC.06478-11. This article demonstrates that the biosynthetic pathway to fosfomycin in Pseudomonas syringae only shares the first and last steps with the pathways characterized in Streptomyces.
- 32.Eliot AC, Griffin BM, Thomas PM, Johannes TW, Kelleher NL, Zhao H, Metcalf WW. Cloning, expression, and biochemical characterization of Streptomyces rubellomurinus genes required for biosynthesis of antimalarial compound FR900098. Chem Biol. 2008;15:765–770. doi: 10.1016/j.chembiol.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johannes TW, DeSieno MA, Griffin BM, Thomas PM, Kelleher NL, Metcalf WW, Zhao H. Deciphering the late biosynthetic steps of antimalarial compound FR-900098. Chem Biol. 2010;17:57–64. doi: 10.1016/j.chembiol.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bae B, Cobb RE, DeSieno MA, Zhao H, Nair SK. New N-acetyltransferase fold in the structure and mechanism of the phosphonate biosynthetic enzyme FrbF. J Biol Chem. 2011;286:36132–36141. doi: 10.1074/jbc.M111.263533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Metcalf WW, Griffin BM, Cicchillo RM, Gao J, Janga SC, Cooke HA, Circello BT, Evans BS, Martens-Habbena W, Stahl DA, et al. Synthesis of Methylphosphonic Acid by Marine Microbes: A Source for Methane in the Aerobic Ocean. Science. 2012;337:1104–1107. doi: 10.1126/science.1219875. This report describes an operon encoding proteins that produce methylphosphonate, and its abundance in metagenomic data sets. These findings could provide an explanation of the previously unknown source of this greenhouse gas in the aerobic ocean waters.
- 36.Karl DM, Beversdorf L, Björkman KM, Church MJ, Martinez A, Delong EF. Aerobic production of methane in the sea. Nat Geoscience. 2008;1:473–478. [Google Scholar]
- 37.Cooke HA, Peck SC, Evans BS, van der Donk WA. Mechanistic Investigation of Methylphosphonate Synthase, a Non-Heme Iron-Dependent Oxygenase. J Am Chem Soc. 2012;134:15660–15663. doi: 10.1021/ja306777w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kamat SS, Williams HJ, Dangott LJ, Chakrabarti M, Raushel FM. The catalytic mechanism for aerobic formation of methane by bacteria. Nature. 2013;497:132–136. doi: 10.1038/nature12061. This groundbreaking work includes reconstitution of several proteins from the Phn operon that catalyze reactions leading to the C-P bond-breaking event. Particularly noteworthy is the reconstitution of the radical SAM enzyme PhnJ responsible for breaking the C-P bond, a feat that had eluded researchers for over 20 years.
- 39.Metcalf WW, Wanner BL. Evidence for a fourteen-gene, phnC to phnP locus for phosphonate metabolism in Escherichia coli. Gene. 1993;129:27–32. doi: 10.1016/0378-1119(93)90692-v. [DOI] [PubMed] [Google Scholar]
- 40.Errey JC, Blanchard JS. Functional annotation and kinetic characterization of PhnO from Salmonella enterica. Biochemistry. 2006;45:3033–3039. doi: 10.1021/bi052297p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Podzelinska K, He SM, Wathier M, Yakunin A, Proudfoot M, Hove-Jensen B, Zechel DL, Jia Z. Structure of PhnP, a phosphodiesterase of the carbon-phosphorus lyase pathway for phosphonate degradation. J Biol Chem. 2009;284:17216–17226. doi: 10.1074/jbc.M808392200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He SM, Wathier M, Podzelinska K, Wong M, McSorley FR, Asfaw A, Hove-Jensen B, Jia Z, Zechel DL. Structure and Mechanism of PhnP, a Phosphodiesterase of the Carbon-Phosphorus Lyase Pathway. Biochemistry. 2011;50:8603–8615. doi: 10.1021/bi2005398. [DOI] [PubMed] [Google Scholar]
- 43.Hove-Jensen B, McSorley FR, Zechel DL. Physiological Role of phnP-specified Phosphoribosyl Cyclic Phosphodiesterase in Catabolism of Organophosphonic Acids by the Carbon-Phosphorus Lyase Pathway. J Am Chem Soc. 2011;133:3617–3624. doi: 10.1021/ja1102713. [DOI] [PubMed] [Google Scholar]
- 44.Hove-Jensen B, Rosenkrantz TJ, Haldimann A, Wanner BL. Escherichia coli phnN, encoding ribose 1,5-bisphosphokinase activity (phosphoribosyl diphosphate forming): dual role in phosphonate degradation and NAD biosynthesis pathways. J Bacteriol. 2003;185:2793–2801. doi: 10.1128/JB.185.9.2793-2801.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jochimsen B, Lolle S, McSorley FR, Nabi M, Stougaard J, Zechel DL, Hove-Jensen B. Five phosphonate operon gene products as components of a multi-subunit complex of the carbon-phosphorus lyase pathway. Proc Natl Acad Sci U S A. 2011;108:11393–11398. doi: 10.1073/pnas.1104922108. This article provides biophysical evidence that Phn operon proteins associate in a larger complex, which set the stage for successful reconstitution of C-P lyase in reference [46].
- 46. Kamat SS, Williams HJ, Raushel FM. Intermediates in the transformation of phosphonates to phosphate by bacteria. Nature. 2011;480:570–573. doi: 10.1038/nature10622. In this mechanistic follow-up paper of the first in vitro reconstitution of C-P lyase, the authors show that catalysis involves amino acid radicals.
- 47.Zhang Q, van der Donk WA. Answers to the Carbon–Phosphorus Lyase Conundrum. Chembiochem. 2012;13:627–629. doi: 10.1002/cbic.201200020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morais MC, Zhang W, Baker AS, Zhang G, Dunaway-Mariano D, Allen KN. The crystal structure of Bacillus cereus phosphonoacetaldehyde hydrolase: insight into catalysis of phosphorus bond cleavage and catalytic diversification within the HAD enzyme superfamily. Biochemistry. 2000;39:10385–10396. doi: 10.1021/bi001171j. [DOI] [PubMed] [Google Scholar]
- 49.Olsen DB, Hepburn TW, Moos M, Mariano PS, Dunaway-Mariano D. Investigation of the Bacillus cereus phosphonoacetaldehyde hydrolase. Evidence for a Schiff base mechanism and sequence analysis of an active-site peptide containing the catalytic lysine residue. Biochemistry. 1988;27:2229–2234. doi: 10.1021/bi00406a063. [DOI] [PubMed] [Google Scholar]
- 50.Szefczyk B. Towards understanding phosphonoacetaldehyde hydrolase: an alternative mechanism involving proton transfer that triggers P-C bond cleavage. Chem Commun. 2008;0:4162–4164. doi: 10.1039/b806280c. [DOI] [PubMed] [Google Scholar]
- 51.Chen CC, Han Y, Niu W, Kulakova AN, Howard A, Quinn JP, Dunaway-Mariano D, Herzberg O. Structure and kinetics of phosphonopyruvate hydrolase from Variovorax sp. Pal2: new insight into the divergence of catalysis within the PEP mutase/isocitrate lyase superfamily. Biochemistry. 2006;45:11491–11504. doi: 10.1021/bi061208l. [DOI] [PubMed] [Google Scholar]
- 52.Cooley NA, Kulakova AN, Villarreal-Chiu JF, Gilbert JA, McGrath JW, Quinn JP. Phosphonoacetate biosynthesis In vitro detection of a novel NADP+−dependent phosphonoacetaldehyde-oxidizing activity in cell-extracts of a marine Roseobacter. Microbiology. 2011;80:335–340. [PubMed] [Google Scholar]
- 53.Borisova SA, Christman HD, Metcalf MEM, Zulkepli NA, Zhang JK, van der Donk WA, Metcalf WW. Genetic and Biochemical Characterization of a Pathway for the Degradation of 2-Aminoethylphosphonate in Sinorhizobium meliloti 1021. J Biol Chem. 2011;286:22283–22290. doi: 10.1074/jbc.M111.237735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim A, Benning MM, OkLee S, Quinn J, Martin BM, Holden HM, Dunaway-Mariano D. Divergence of chemical function in the alkaline phosphatase superfamily: structure and mechanism of the P-C bond cleaving enzyme phosphonoacetate hydrolase. Biochemistry. 2011;50:3481–3494. doi: 10.1021/bi200165h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Agarwal VSA, Borisova SA, Metcalf WW, van der Donk WA, Nair SK. Structural and Mechanistic Insights into C-P Bond Hydrolysis by Phosphonoacetate Hydrolase. Chem Biol. 2011;18:1230–1240. doi: 10.1016/j.chembiol.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gerlt JA, Babbitt PC, Rayment I. Divergent evolution in the enolase superfamily: the interplay of mechanism and specificity. Arch Biochem Biophys. 2005;433:59–70. doi: 10.1016/j.abb.2004.07.034. [DOI] [PubMed] [Google Scholar]
- 57. Martinez A, Tyson GW, DeLong EF. Widespread known and novel phosphonate utilization pathways in marine bacteria revealed by functional screening and metagenomic analyses. Environ Microbiol. 2010;12:222–238. doi: 10.1111/j.1462-2920.2009.02062.x. In addition to the better-known hydrolytic and reductive routes to breakdown of phosphonates, this report provides the first evidence that organisms also utilize oxidative catabolic routes. This study also demonstrates a strategy to find new catabolic pathways.
- 58.McSorley FR, Wyatt PB, Martinez A, DeLong EF, Hove-Jensen B, Zechel DL. PhnY and PhnZ Comprise a New Oxidative Pathway for Enzymatic Cleavage of a Carbon-Phosphorus Bond. J Am Chem Soc. 2012;134:8364–8367. doi: 10.1021/ja302072f. [DOI] [PubMed] [Google Scholar]
- 59.Bollinger JM, Jr, Diao Y, Matthews ML, Xing G, Krebs C. myo-Inositol oxygenase:a radical new pathway for O2 and C-H activation at a nonheme diiron cluster. Dalton Trans. 2009:905–914. doi: 10.1039/b811885j. [DOI] [PMC free article] [PubMed] [Google Scholar]