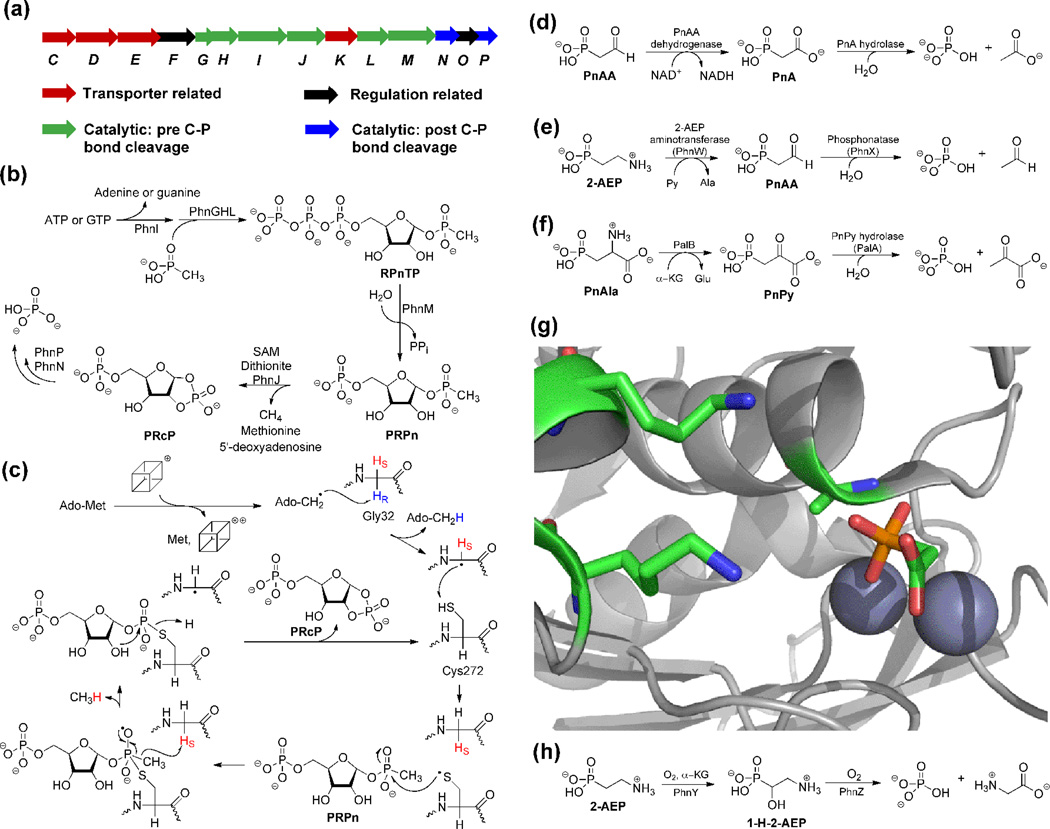
Figure 4.
Overview of phosphonate catabolism. (a) Gene cluster of the Phn operon for reductive C-P bond cleavage. (b) Schematic overview of the C-P lyase pathway. (c) A recently proposed mechanism for PhnJ catalyzed C-P bond cleavage [38]. The cube represents the [4Fe-4S] cluster. (d) Phosphonoacetate hydrolase (PhnA) is a newly discovered hydrolase that takes advantage of the carboxylate group as a "β electron sink". (e) Phosphonatase is able to convert 2-AEP to free phosphate through PnAA. (f) PnPy hydrolase is used to liberate phosphate from PnPy after transamination of PnAla. (g) The two lysines (shown as sticks) in the PhnA-T68A active site are relatively far from PnA. Ala68 is set up for in-line attack on the phosphonate when this residue is Thr in wild type PhnA, and the π-system of the carboxylate of PnA is well oriented to accept electron density upon cleavage of the P-C bond. The incipient negative charge may also be stabilized by binding to a Zn(II) in a monodentate fashion. (h) A recently discovered pathway from a genomic marine library results in oxidative cleavage of the C-P bond.