Figure 1.
Erk1/2 and Gsk3β Signal Inhibition Induces Global DNA Demethylation
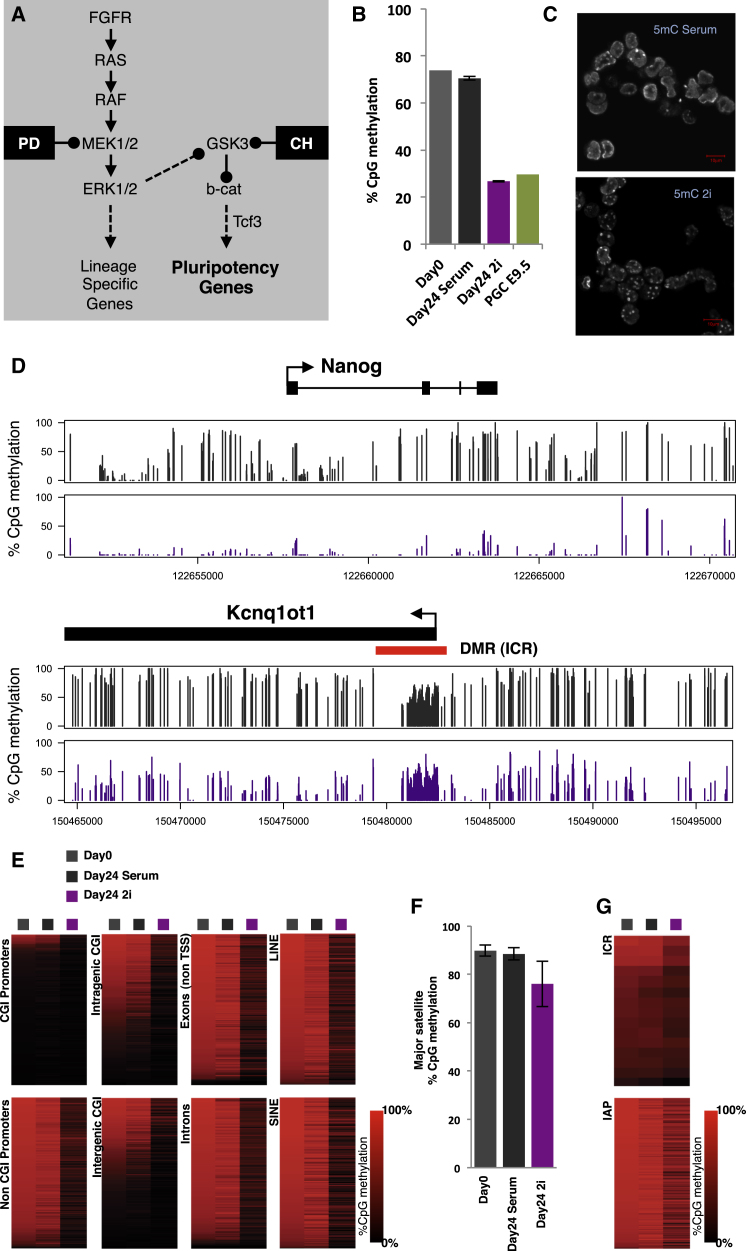
(A) Schematic of the signaling pathways inhibited by the “2i” small molecule inhibitors.
(B) Global CpG methylation measured by whole-genome BS-seq in serum, 2i, and E9.5 PGCs (data from Seisenberger et al., 2012). Error bars represent the standard deviation in three replicates.
(C) Immunofluorescence staining of E14 ESCs with an antibody against 5mC shows reduced euchromatic methylation in 2i while pericentromeric heterochromatic regions maintain high 5mC levels.
(D) Example of BS-seq profile in serum (black bars) and 2i (purple bars) ESCs with the Nanog locus being strongly demethylated in 2i while the ICR methylation at Kcnq1ot1 is maintained.
(E) Heatmap methylation levels in 500 randomly selected elements (CpG islands (CGIs), Exons, Introns, LINE1, SINE) in Day 0, Day 24 Serum, and Day 24 2i.
(F) Confirming IF data, methylation at pericentromeric major satellites remains high in 2i as measured by BS-seq (error bars represent the standard deviation between CpGs).
(G) Heatmap methylation levels in 500 randomly selected IAP elements and 15 ICRs.
See also Figure S1.