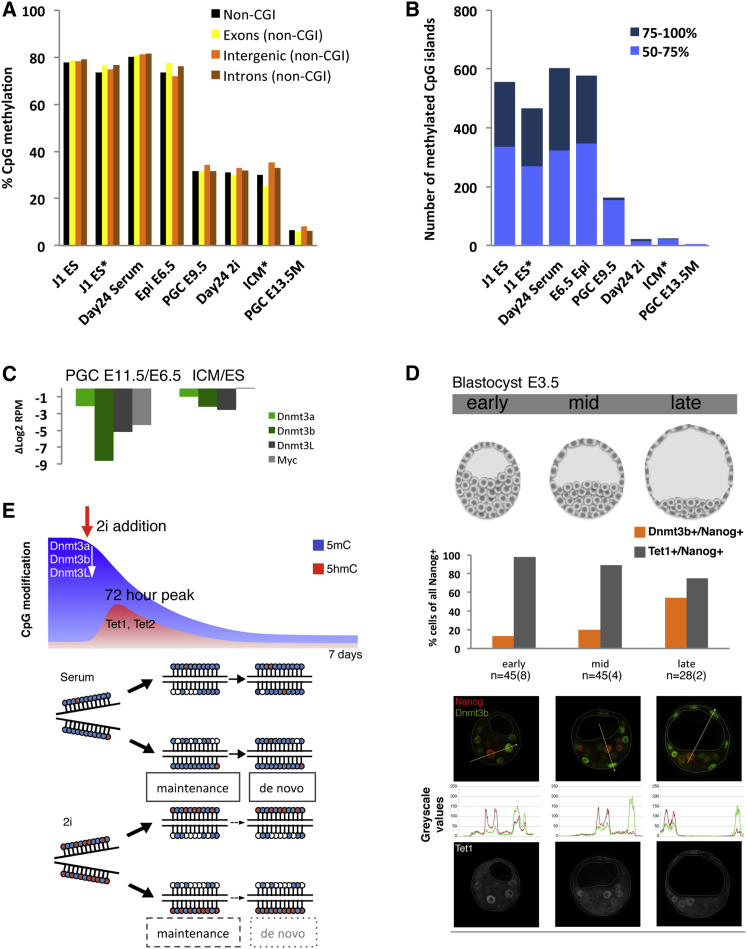
Figure 4.
Global Hypomethylation in 2i ESCs Resembles that of ICM Cells and Migratory PGCs
(A) Global methylation levels in ESCs, ICM cells, and PGCs from whole-genome BS-seq data and reduced representation BS-seq (RRBS) data sets (RRBS data are labeled with asterisk) (Seisenberger et al., 2012; Smith et al., 2012).
(B) Number of highly methylated (75%–100%) or moderately methylated (50%–75%) CGIs of the same samples in (A).
(C) Expression of de novo methyltransferases Dnmt3a, Dnmt3b, Dnmt3L, and Myc in PGC E11.5 compared to E6.5 epiblast cells and ICM versus ESCs (Tang et al., 2010; Seisenberger et al., 2012).
(D) E3.5 blastocysts stained for NANOG (red), DNMT3B (green), and TET1 (white). Mouse E3.5 blastocysts were collected from CD1 × CD1 and (C57BL/6 × CBA/Ca) × 129/Sv crosses and were classified as early, mid, or late E3.5, depending on their size and extent of cavitation. Late blastocysts show higher DNMT3B expression as well as a greater proportion of ICM cells expressing both NANOG and DNMT3B and slightly fewer NANOG- and TET1-expressing cells (the number of Nanog-positive cells counted from the total number of embryos in parenthesis is shown below the barplot). RGB profiles for representative ICM (NANOG high) and outer trophectoderm (TE) cells are shown for each stage.
(E) Model for 2i-mediated demethylation including a peak of oxidation and reduced efficiency of maintenance methylation and disruption of de novo methylation.