Abstract
In his original exposition of the biogenetic isoprenoid rule, L. Ruzicka noted the structural identity between the fused A/B rings of triterpenoids/sterols and certain multicyclic diterpenoids as part of the impetus leading to that profound insight. His prescient hypothesis that this chemical structure relationship reflects similarities in the initial cyclization of these diterpenoids with that occurring in triterpenoid biosynthesis has since been verified. However, this chemical structure relationship does not continue to hold true for the additional rings found in many of these di- and tri- terpenoid natural products. This is now appreciated to arise from differences in their subsequent biogenesis, specifically further cyclization and/or rearrangement of these diterpenoids after formation of an initial bicyclic intermediate in a separately catalyzed reaction. The trivial name for the hydrocarbon skeleton of the most commonly found version of the corresponding unique intermediate forms the basis for a unifying “labdane-related” designation. This defines a large super-family of diterpenoids that contains nearly 7,000 already known natural products. Many of these are found in plants, where the requisite biosynthetic machinery for gibberellin phytohormones, particularly the relevant diterpene cyclases, provides a biosynthetic reservoir that appears to have been repeatedly drawn upon to evolve new labdane-related diterpenoids. The potent biological activity of the “ancestral” gibberellins, which has led to the independent evolution of distinct gibberellin biosynthetic pathways in plants, fungi, and bacteria, is further discussed as an archetypical example of the selective pressure driving the observed diversification of the large super-family of labdane-related diterpenoid natural products.
1 Introduction
Natural products are generally classified on the basis of their biogenetic origins, leading to the typical terpenoid/isoprenoid, phenylpropanoid, and alkaloid classifications, with the constituent compounds further divided on the basis on their more specific origins; e.g., diterpenoids, flavonoids, and purine alkaloids.1 Of particular interest here are the diterpenoids, derived from (E,E,E)-geranylgeranyl diphosphate (GGPP), which form a large clan of >12,000 natural products.2 Notably, the diterpenoids can be further divided on the basis of biogenetic hydrocarbon ring construction, with a significant fraction of the polycyclic diterpenoids (~7,000) arising from dual, rather than single, biosynthetic cyclization and/or rearrangement reactions, regardless of final number of rings. Here a unifying labdane-related diterpenoid designation is proposed for these to reflect their distinct biogenetic origins.
2 Unifying biogenetic origins of labdane-related diterpenoids
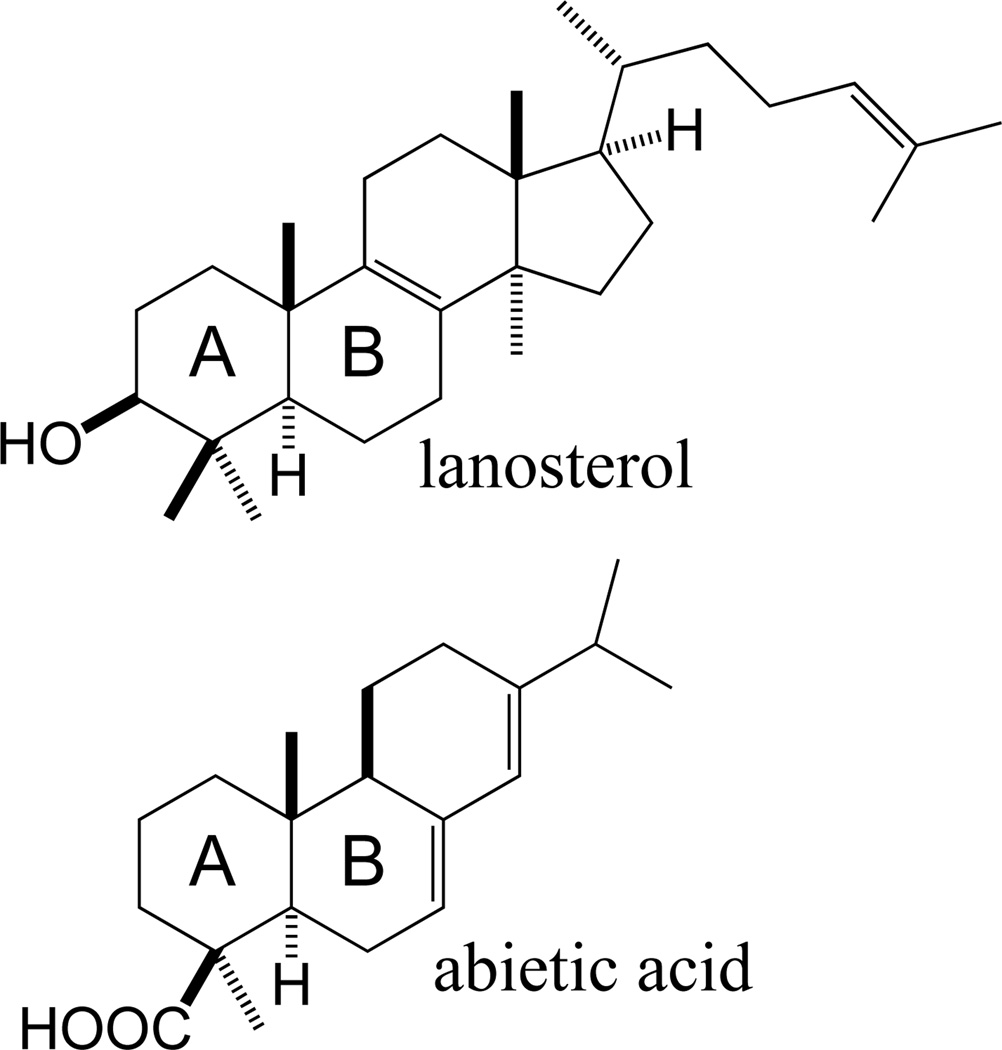
The identical stereochemical configuration of the bicyclic decalin ring structure at the core of triterpenoid and certain diterpenoid natural products, specifically the trans nature of the substituents on the bridging carbons (Figure 1), was noted as part of the impetus behind development of the “biogenetic isoprene rule”.3 This was a prescient hypothesis that has since been fully supported, including evidence for analogous cyclization mechanisms leading to the decalin core ring structure.4 Specifically, the decalin ring structure is now known to be formed by a protonation-initiated, cationic cycloisomerization reaction in both cases, while the differences in additional ring structure are further recognized as indicative of their distinct biogenetic origins in the further cyclization of di- and tri- terpenoid natural products. In particular, it has become apparent that this difference in subsequent ring structure arises from the continuing nature of triterpenoid cyclization, while that of the relevant polycyclic diterpenoids is carried out by two distinct mechanisms, as first suggested by C. A. West.5 Initial cyclization is mediated by the aforementioned protonation-initiated reaction, which most commonly results in a hydrocarbon skeletal structure analogous to the labdanes, albeit with the diphosphate group remaining from the GGPP precursor, forming a labdadienyl/copalyl diphosphate (CPP) intermediate. Accordingly, although not all natural products sharing such dual cyclization reaction biogenetic origins are produced via this particular intermediate, the labdane-related diterpenoid nomenclature presented here stems from this distinctive structure. In the remainder of this section, the unifying biogenetic origins of this super-family of natural product will be discussed in more detail. Given the recent advances in this area, the characterization and/or identification of relevant enzymes is specifically included here.
Figure 1.
2.1 Initiating class II diterpene (bi)cyclization
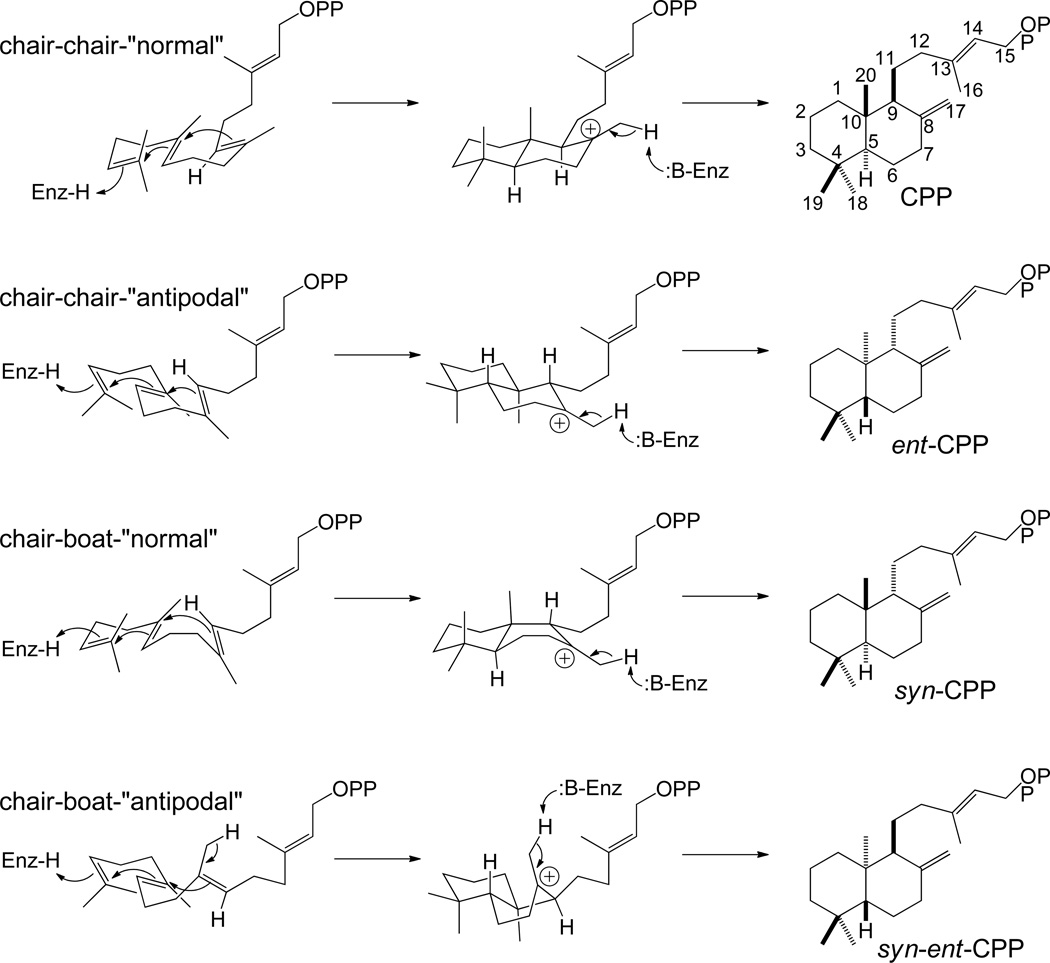
The initiating and defining step in labdane-related diterpenoid biogenesis is the protonation-initiated cyclization of GGPP, which proceeds via cationic carbon-carbon double bond addition.6,7 Notably, although mono- or tri-cyclization does appear to be possible, there do not appear to be any examples of corresponding natural products that are unambiguously derived from such class II cyclization of GGPP rather than being degradative products of tri- or tetra- terpenoids. Specifically, the monocyclic retinanes could be derived from carotenoid-like tetraterpenes, and the tricyclic spongianes are likely degraded triterpenoid quassinoids.8 Accordingly, this reaction has only been definitively shown to mediate bicyclization of GGPP to a labda-13-en-8-yl+ diphosphate intermediate. The stereochemistry of this intermediate depends on the pro-chiral conformation of the GGPP precursor upon catalysis, albeit with the fixed trans configuration across the decalin bridgehead noted by Ruzika that is imposed by the anti-parallel addition cyclization mechanism. In particular, a pro-chair-chair conformation leads to copalyl of either normal, designated by comparison to the stereochemistry of the analogous A/B rings in cholesterol,8 or antipodal/enantiomeric (ent) stereochemistry, depending on absolute configuration, while a pro-chair-boat conformation leads to syn orientation of the methyl and hydride substituents across the C-9,10 bond, which can occur with either the normal or ent absolute configuration. These various stereoisomers are most commonly quenched by deprotonation of the methyl group at C-8, forming the exo-methylene of the previously noted CPP, whose stereochemistry is then designated as normal, ent, syn, or syn-ent, respectively (Scheme 1).9 Notably, the most commonly observed variant is ent-CPP, with many such class II diterpene cyclases known,10–25 particularly from plants where they are required for gibberellin phytohormone metabolism (as discussed in Section 4). Normal CPP also is quite common, with several such enzymes known,26–31 while syn-CPP is much less common and only a few such enzymes are known.32 By contrast, the production of syn-ent-CPP has not been observed, and only natural products from plants of the Calceolaria genus have been rigorously demonstrated to have such stereochemistry.33 Notably, the defining stereochemistry at C-9 and C-10 of the corresponding CPP often remains unchanged in the extended families of derived labdane-related diterpenoid natural products, enabling assignment of the corresponding biosynthetic intermediate with some confidence.
Scheme 1.
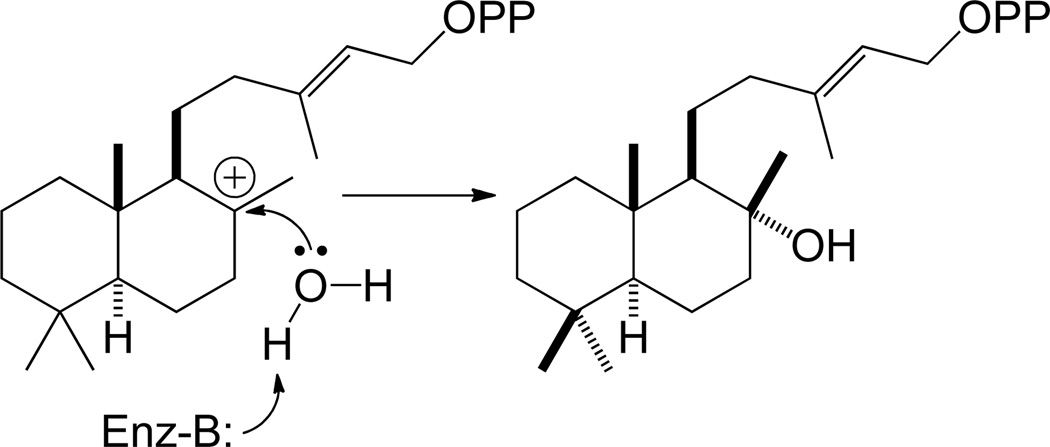
Beyond simply generating CPP by the cycloisomerization reaction described above, class II diterpene cyclases also are capable of generating further chemical diversity. In particular, it is possible for the initially bicyclized labda-13-en-8-yl+ diphosphate carbocation to be captured by water prior to deprotonation, leading to hydroxylated variants (labda-13-en-8-ol diphosphate with the hydroxyl in either the α or β position). Presumably, such quenching is most readily achieved by anti-parallel addition of the water, such that hydroxyl group stereochemistry is at least biased, if not dictated, by GGPP pro-chiral configuration. Indeed, class II diterpene cyclases catalyzing this type of cyclohydration produce a compound with just such stereochemistry, specifically labda-13-en-8α-ol diphosphate of normal configuration (Scheme 2).34
Scheme 2.
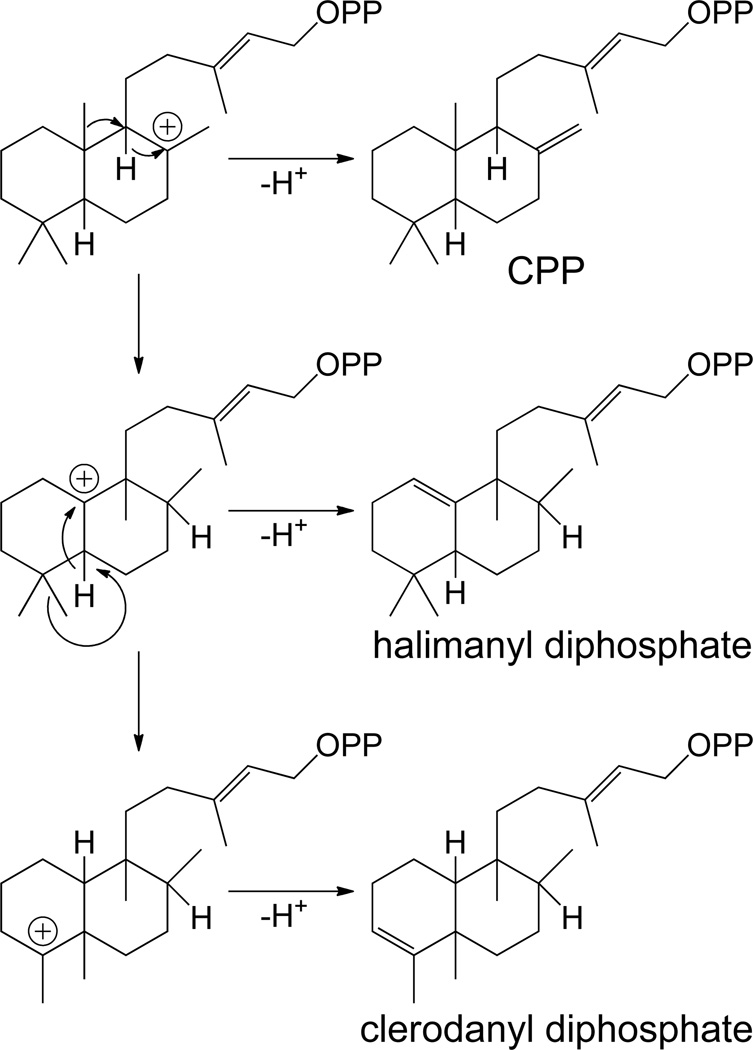
This class II (bi)cyclization reaction also is capable of generating further hydrocarbon skeletal diversity, as the initially formed labda-13-en-8-yl+ diphosphate intermediate can undergo a series of 1,2-hydride and/or methyl shifts, much like those observed in the analogous triterpenoid cyclization reactions,35 with the full series forming the clerodane hydrocarbon backbone. These migrations do not appear to be concerted, as the only identified class II diterpene cyclase that produces a clerodadienyl/terpentedienyl diphosphate contains an anti arrangement of methyl substituents on C-8 and C-9 that must arise from syn hydride/methyl migration.36 In addition, the last methyl shift also does not appear to be restricted to anti-parallel migration, as cis and trans arrangements of the substituents on the bridging C-5/C-10 are known in natural products from the clerodane family, with production of the alternative cis bridgehead arrangement within the class II cyclization reaction enabled by the presence of geminal methyl groups on C-4, which allows alternative migrations dictating the configuration of the resulting bridgehead. Furthermore, this series of shifts can be interrupted at various stages by quenching of the relevant carbocationic intermediate, most commonly by deprotonation of a neighboring endo-methylene, leading to a series of rearranged isoprenyl diene diphosphate compounds (Scheme 3), albeit with retention of the bicyclic decalin core. The most easily recognized variant is that corresponding to the halimadane backbone, resulting from single hydride and methyl migrations. The double bond isomers of halimadienyl diphosphate can be generated by deprotonation either before or after hydride migration from C-5 to C-10, and a bacterial class II diterpene cyclase that produces halima-5,13-dienyl diphosphate (i.e. post-C5,10 hydride shift) has been identified.37
Scheme 3.
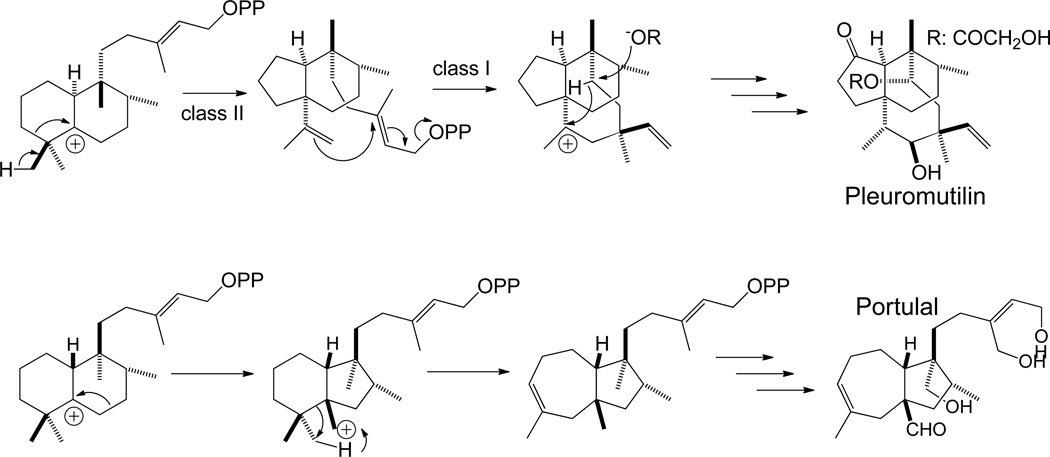
Intriguingly, there is strong evidence for B-ring contraction proceeding from the preceding haliman-13-en-5-yl+ diphosphate intermediate in plueromutulin biosynthesis,38 providing an example of how this reaction may be used to generate even more diverse hydrocarbon backbone structures (Scheme 4). Indeed, several alternative such ring rearrangements may be catalyzed by class II diterpene cyclases (e.g. in portulal biogenesis). Finally, while it should be possible to generate further chemical diversity by capture of any of these carbocation intermediates with water prior to deprotonation (much as described above), it does not appear that any such class II diterpene cyclases have been characterized.
Scheme 4.
2.2 Subsequent class I diterpene synthase transformations
Class II cyclization reactions leave intact in the resulting bicyclic isoprenyl the allylic diphosphate ester linkage from the GGPP precursor. This is then utilized by terpene synthases analogous to those operating in terpenoid biosynthesis more generally,39 to catalyze lysis/ionization-initiated, cationic cyclization and/or rearrangement reactions. Notably, the terpene synthases operating in labdane-related diterpenoid biosynthesis specifically act upon the bicyclic isoprenyl diphosphate precursors formed by the upstream class II diterpene cyclases. In particular, these class I diterpene synthases are generally much less reactive with the more universal diterpenoid precursor GGPP, and other acyclic isoprenyl diphosphates, although they may react with a range of class II diterpene cyclase products (e.g., two different stereoisomers of CPP).40 The hydrocarbon backbone structure formed by these class I diterpene synthases typically remains largely unchanged in the derived natural products, providing the basis for their assignment into families of biogenetically related natural products. A number of such skeletal structures are described in Section 3 below, and the corresponding class I reaction mechanisms will be discussed there. Here it is simply noted that there are a significant number of such backbone structures known, particularly when differences in sterochemical configuration are further taken into account. Finally, it should also be noted that the various carbocation intermediates in these class I reactions also can be captured by water prior to deprotonation, just as described above for the class II reactions, with the additional possibility of (re)capture of the ionized pyrophosphate anion, as described for the monoterpene cyclase bornyl diphosphate synthase,41 although no other terpenoid has yet been characterized as having an analogous biogenetic origin.
2.3 Further elaboration
Much of the diversity in any group of terpenoid natural products arises from the tailoring reactions that decorate the basic hydrocarbon backbone,1 and the labdane-related diterpenoids are no exception. Accordingly, while the primary focus of this review is the unifying nature of their biogenetic origins, the range and type of downstream modifications will be briefly mentioned here. Such modification almost invariably is initiated by the insertion of oxygen catalyzed by a cytochrome P450 monooxygenase. The resulting hydroxyl then provides a target for further transformations, most generally, addition of functional groups. These range from a simple methyl or acetate group to amino acids, sugars, and fatty acids, with the labdane-related diterpenoids able to further form part of more complex (meroterpenoid) natural products, such as the indole diterpenoid alkaloids.2 The appended functional groups can be targeted for further transformations and/or addition, providing even more, albeit auxiliary chemical diversity. Ultimately, it is these tailoring reactions that produce the wide range of observed labdane-related natural products, with differences in resulting structure defining the ~7,000 individual members of this super-family. It also should be noted here that these downstream transformations can lead to rearrangement of the hydrocarbon backbone structure formed by the initiating dual cyclization reactions described above, obscuring assignment of the resulting natural product(s) to a particular family, although this can, and in many cases has been worked out, as will be described for the ent-kaurene derived gibberellin phytohormone and oryzalide phytoalexin families below (Section 3.5).
3 Prototypical labdane-related diterpenoids
To provide an overview of the labdane-related diterpenoids, and particularly the unifying biogenetic origins of the hydrocarbon backbone that defines the constituent families of natural products, a number of the most common are presented here, along with the relevant class I cyclization reaction mechanisms and, where known, the corresponding enzymes.
3.1 Bicyclic: Labdanes, clerodanes, and others
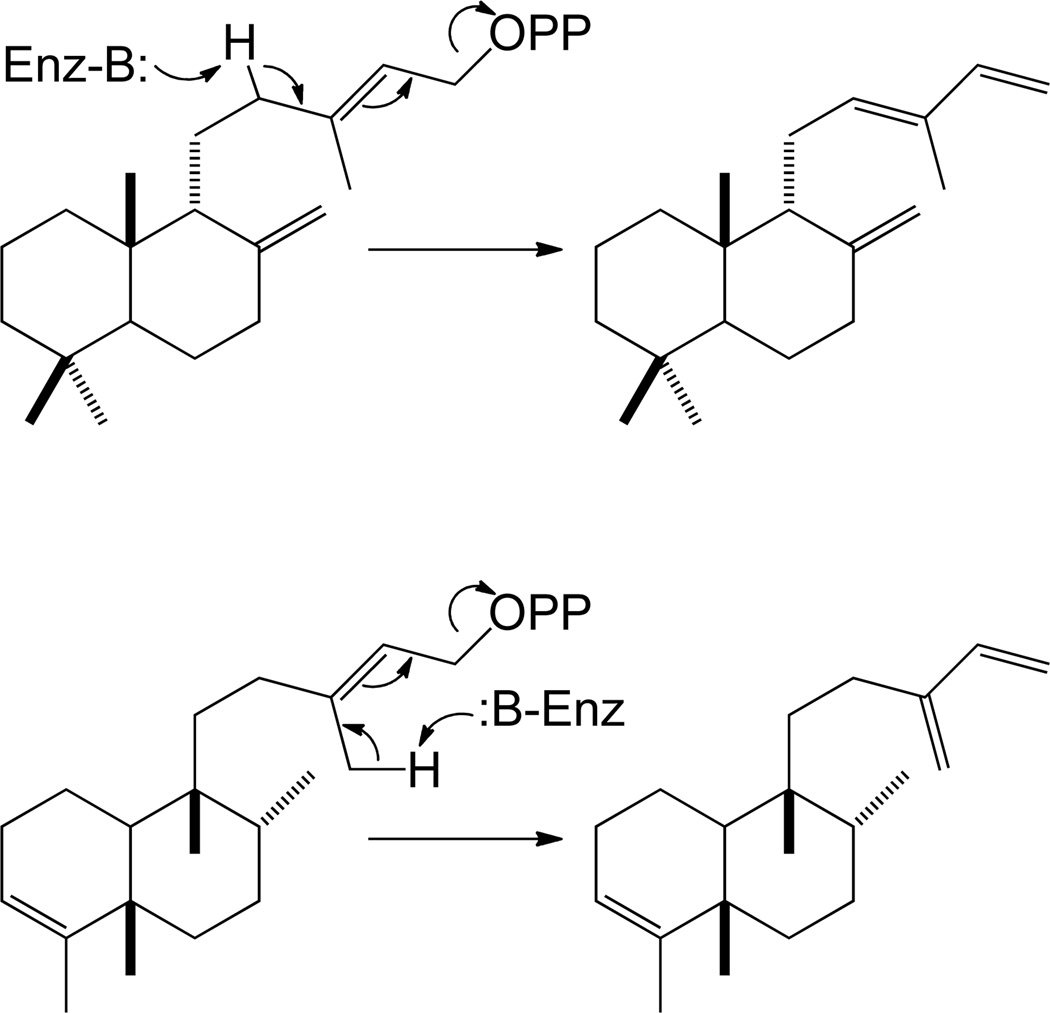
The most basic hydrocarbon backbones in the labdane-related diterpenoid super-family are the bicyclic such natural products, which include the labdane, clerodane and halimadane families. As described above, the underlying ring structure is formed by the relevant class II diterpene cyclase, with the relevant class I diterpene synthases presumably then simply removing the diphosphate without catalyzing cyclization. Most typically, it would be expected that this results in an olefin via direct deprotonation of the initially formed allylic cation, typically at the neighboring endo-methylene or methyl. For example, enzymes forming syn-labda-8(17),12E,14-triene and clerodatriene/terpente-3,13(16),14-triene have been identified (Scheme 5).36, 40 In addition, just as described for the class II diterpene cyclases, it is possible for this intermediate to be captured by water prior to deprotonation, yielding a hydroxylated diterpenoid, and a class I diterpene synthase catalyzing just such a reaction has been very recently identified (personal communication; M. Schalk, Firmenich). Notably, while further cyclization of rearranged class II bicycles seems possible (e.g. with halimadienyl diphosphate), this has only been specifically invoked in the case of pleuromutilin biosynthesis (Scheme 4). Thus, the overwhelming majority of further cyclized labdane-related diteprenoids are derived from the various stereoisomers of CPP.
Scheme 5.
3.2 Tricyclic: Pimaranes, abietanes, and others
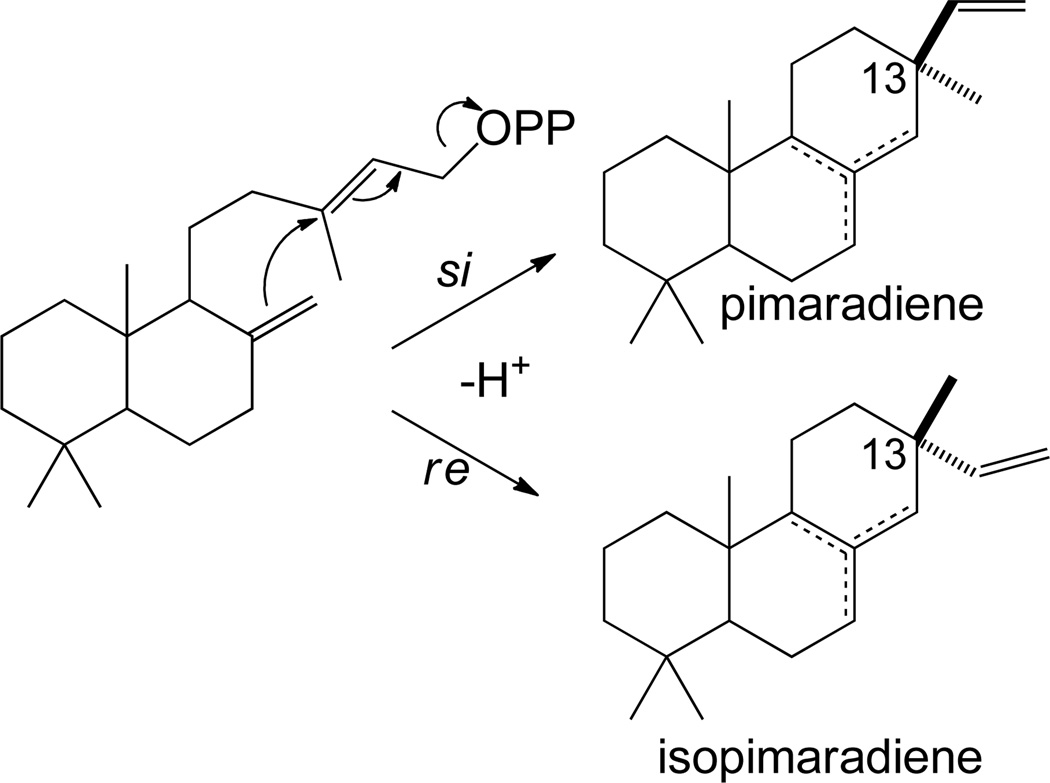
Given that further cyclization seem to almost invariably proceed from CPP, attack of the initially formed allylic carbocation on the C-8(17) carbon-carbon double bond inevitably occurs via Markovnikov addition, reflecting the much more favorable formation of a tertiary versus primary carbocation. This can occur via either si-face or re-face attack/addition, leading to alternative configurations of the geminal methyl/vinyl pair at C-13 of the resulting pimara-15-en-8-yl+ intermediate,9 with the α-methyl/β-vinyl configuration considered normal (which seems to be based on early studies of resin acid configuration) and the inverse β-methyl/α-vinyl the epi/isomeric form. Direct deprotonation of this carbocationic intermediate from any of the neighboring endo-methylenes yields various double bond isomers of (iso)pimaradiene (Scheme 6). Class I diterpene synthases catalyzing the production of pimaradienes or isopimaradienes from normal, ent-, and syn-CPP have been identified.28, 40, 42–45
Scheme 6.
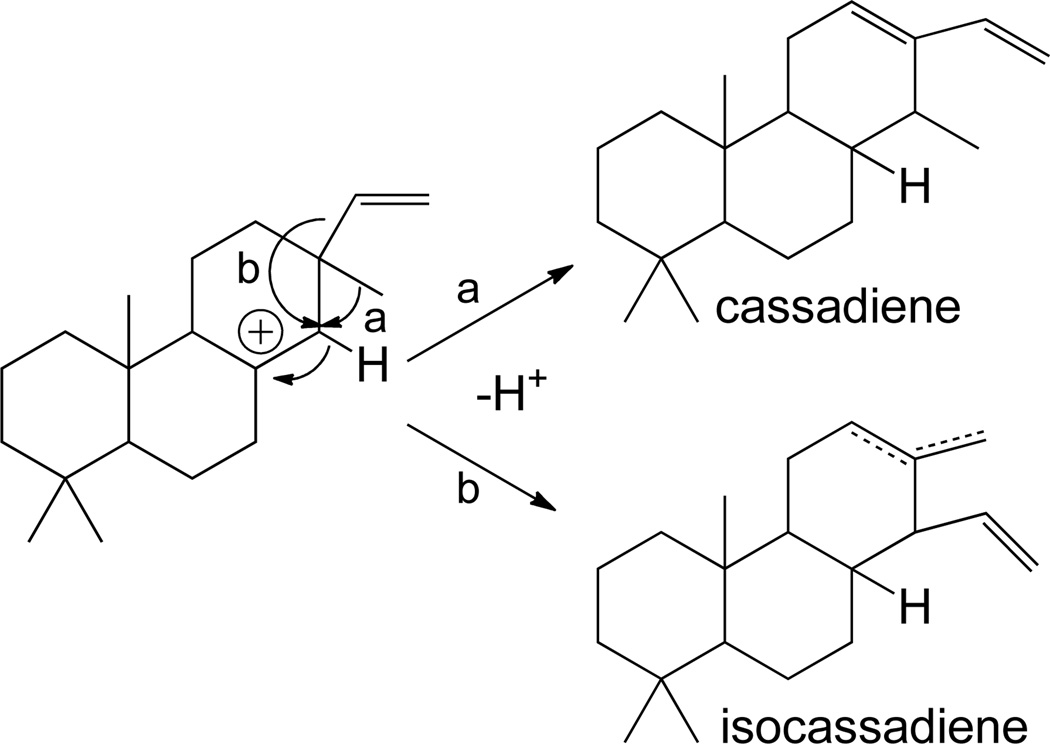
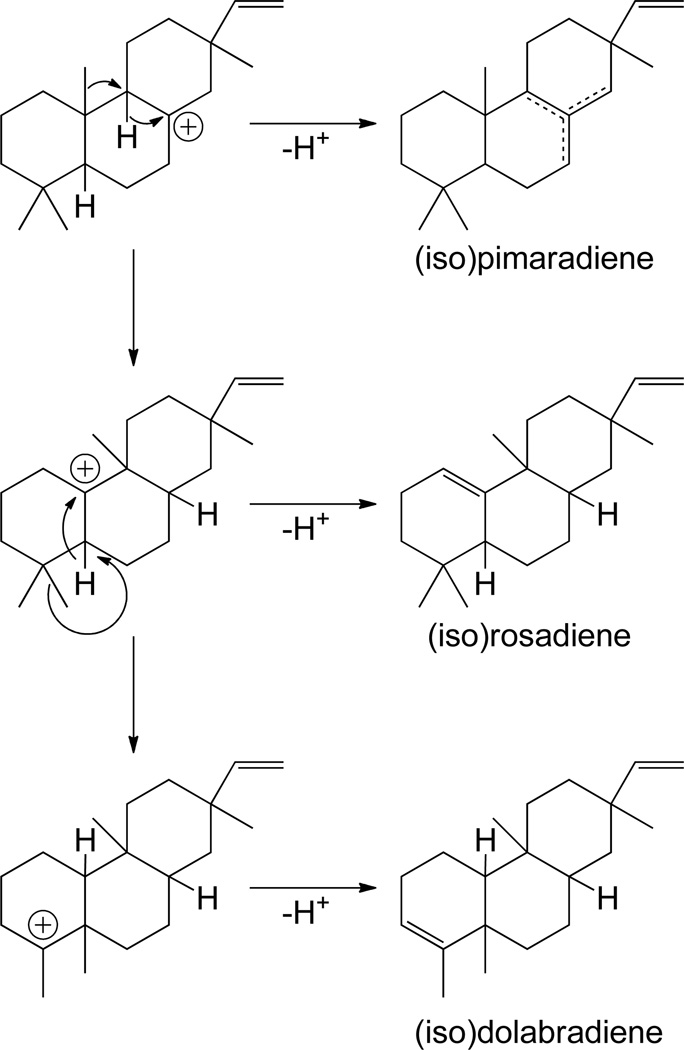
Even beyond these various (iso)pimaradienes, other tricycles are known, arising from rearrangement of the initially (tri)cyclized (iso)pimara-15-en-8-yl+ intermediate prior to quenching of a terminal carbocation.8, 9 The most trivial example arises from a 1,2-hydride shift, followed by deprotonation of a neighboring endo-methylene, yielding a 9(11)-double bond isomer of (iso)pimaradiene, with one such class I diterpene synthase already identified.22 More interestingly, the various corresponding carbocation intermediates can undergo further rearrangements. The pimara-15-en-14-yl+ intermediate resulting from C14,8 hydride transfer can lead to a 1,2-shift of either substituent from the neighboring C-13, with methyl migration forming cassa-15-en-13-yl+ and vinyl migration isocassa-15-en-13-yl+ intermediates, followed by direct deprotonation to form the corresponding diene, and an ent-cassa-12,15-diene synthase has been identified (Scheme 7).46 Notably, C9,8 hydride transfer can initiate a series of 1,2 methyl and hydride migrations analogous to the rearrangements discussed for class II reactions above (i.e., those leading to clerodanes; Scheme 8). Methyl migration from C-10 to C-9 forms a rosa-15-en-10-yl+ intermediate, with ensuing 1,2 hydride transfer leading to a rosa-15-en-5-yl+ intermediate, either of which can be deprotonated on a neighboring endo-methylene to form the corresponding diene. Finally, the latter intermediate can undergo a 1,2 methyl transfer to form a dolabra-15-en-4-yl+ intermediate that can either be directly deprotonated to the corresponding diene, or form a cyclopropyl ring by deprotonation of the delocalized methyl in the process of migrating, yielding the tetracycle devadarene. However, it must be noted that no enzymes catalyzing any of the 1,2 methyl/hydride shift mediated rearrangements discussed above appear to have been characterized to date.
Scheme 7.
Scheme 8.
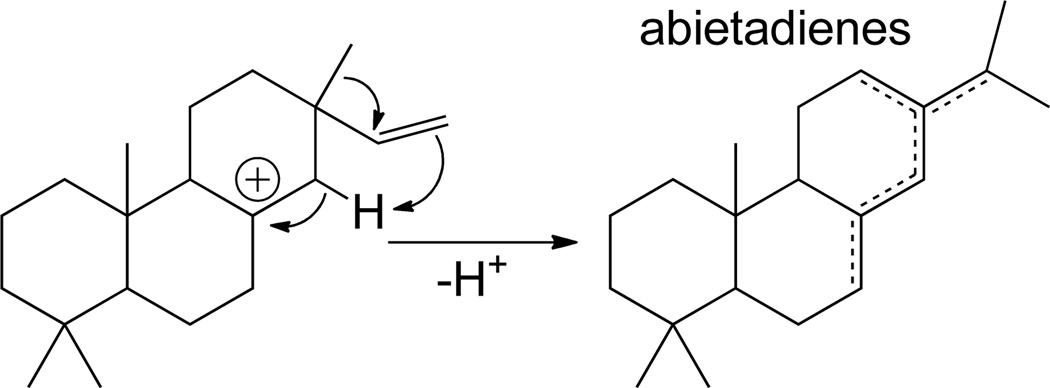
By contrast, it has been shown that the initially formed pimara-15-en-8-yl+ intermediate can undergo a 1,4 proton transfer (C-14,16) to produce a pimara-8(14)-en-15-yl+ intermediate that can undergo rearrangement.47 In particular, a 1,2 methyl migration to form abieta-8(14)-en-13-yl+, which can be deprotonated in variety of positions to form the relevant dienes (Scheme 9).48 Several such class I diterpene synthases have been indentified,26, 28, 29 including one suggested to catalyze 1,5 proton transfer in order to produce abieta-8,12-diene (miltiradiene).31 Alternatively, the C-ring of the pimara-8(14)-en-15-yl+ intermediate can undergo ring expansion and deprotonation of the neighboring endo-methylene to form stroba-8(15),12-diene, although it does not appear that an enzyme catalyzing such a reaction has yet been characterized.
Scheme 9.
3.3 Tetracyclic: Kauranes, beyeranes, and others
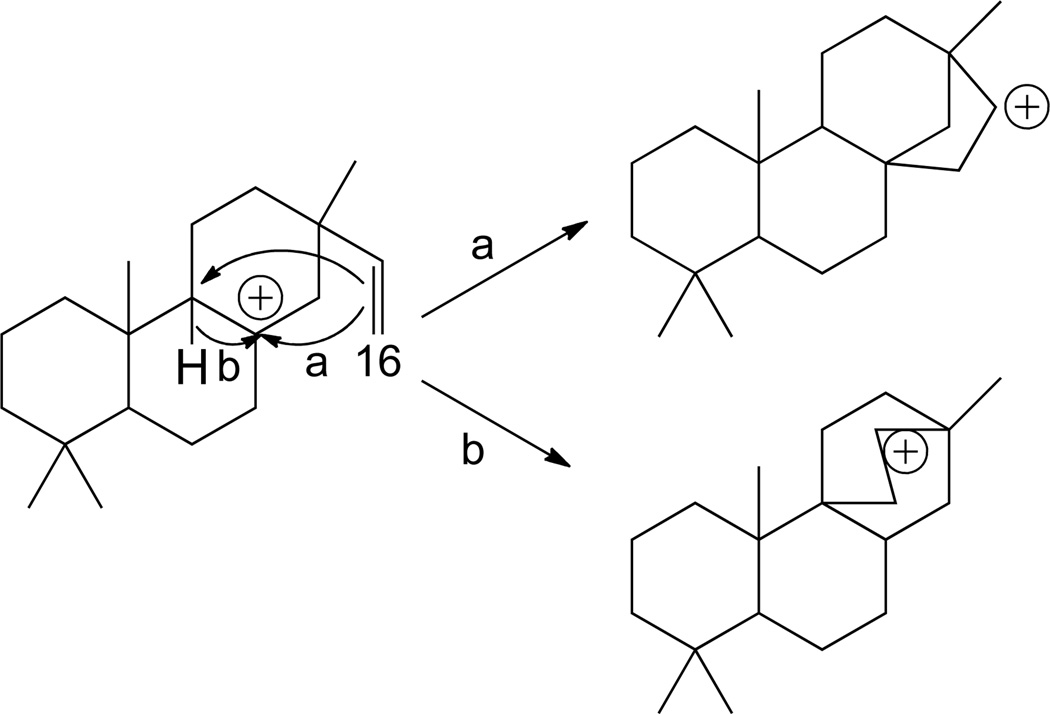
Given that tricyclization generally occurs from CPP to form (iso)pimarenyl+ intermediates, tetracycles are then formed by attack of the terminal C-16 methylene of the C-13 vinyl group on either C-8 or, following C-9,8 hydride transfer, C-9 (Scheme 10).9 Given the potential for four different stereoisomers of CPP, and potential si-face or re-face tricyclization to eight different variants of (iso)pimarenyl+, there are then sixteen different potential tetracycles. In addition, these tetracyclic intermediates can then undergo additional rearrangement, potentially leading to at least 75 different skeletal structures, although only some of these have been actually shown to occur. Here only several examples will be presented in an attempt to provide an overview of the potential types of tetracycles, with a focus on those with known biogenetic relevance.
Scheme 10.
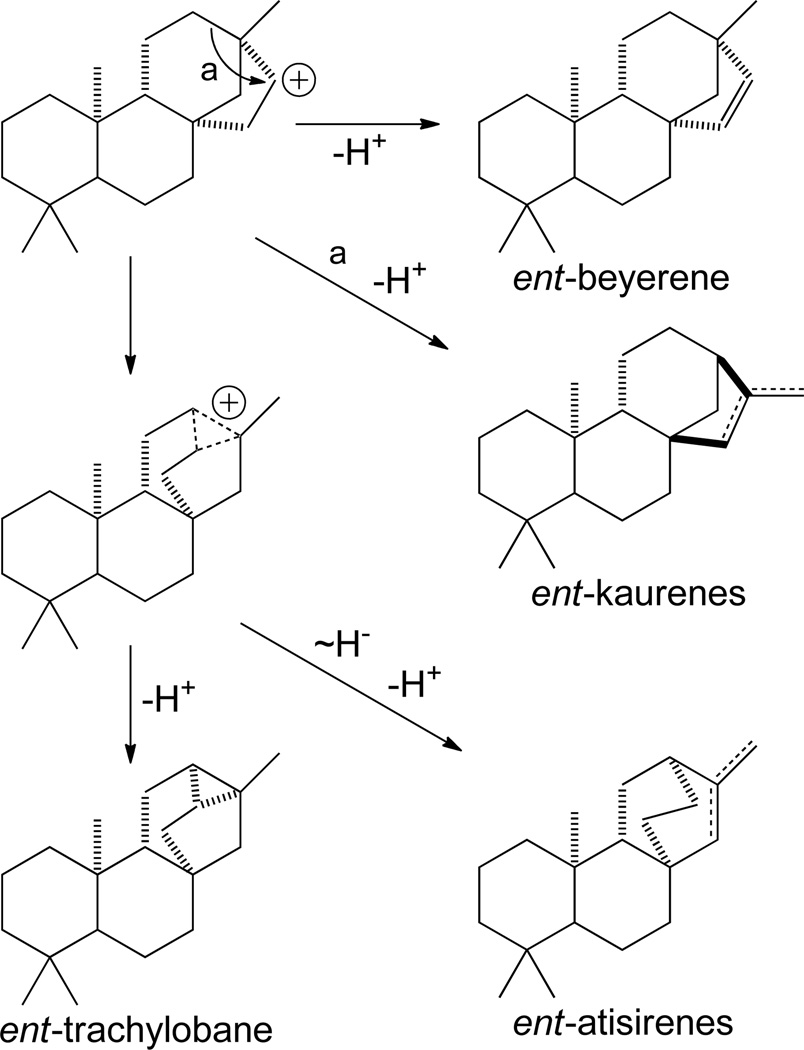
Starting from the ent-pimara-15-en-8-yl+ intermediate arising from re-face tricyclization of ent-CPP, tetracyclization connecting C-16 to C-8 forms a ent-beyer-15-yl+ intermediate, which can be directly deprotonated on the neighboring endo-methylene to form ent-beyer-15-ene. Alternatively, the secondary carbocation ent-beyer-15-yl+ intermediate can undergo ring rearrangement (Scheme 11). Intriguingly, this proceeds via a delocalized carbocationic intermediate, which can be directly deprotonated to the pentacycle trachylobane, or yield tertiary carbocation intermediates by either undergoing a 1,2 hydride shift and ring arrangement to form ent-atisir-16-yl+ or more straightforward single bond shift to form ent-kaur-16-yl+. In both of the latter two cases, these tertiary carbocations can be directly deprotonated at either the neighboring methyl or endo-methylene, to yield the corresponding double bond isomers. Of particular interest, deprotonation of ent-kaur-16-yl+ on the neighboring methyl group gives rise to the ent-kaur-16-ene relevant to gibberellin phytohormone biosynthesis, while deprotonation of the neighboring endo-methylene yields ent-isokaur-15-ene. Given the role of ent-kaurene synthases in gibberellin phytohormone metabolism, it is present in all higher plants, the implications of which are described below (Section 4), and many such enzymes have already been identified.19, 21, 24, 25, 45, 49–51 Notably, these selectively produce ent-kaur-16-ene, while selective ent-isokaur-15-ene synthases also have been identified.44, 45 Furthermore, it is possible to add water prior to deprotontation, to form ent-kaur-16-ol, and a diterpene synthase catalyzing such product outcome has been identified as well.52 In addition, a mutant ent-pimaradiene synthase has been found to produce some ent-atisir-16-ene,53 with both ent-beyerene and trachylobane synthase activity having been characterized in castor bean.54 An equivalent series of transformations is known for cyclization of normal CPP, with pimar-15-en-8-yl+ derived kaur-16-ene known, and a diterpene synthase producing the isopimar-15-en-8-yl+ derived phyllocladan-16-ol has been identified.30
Scheme 11.
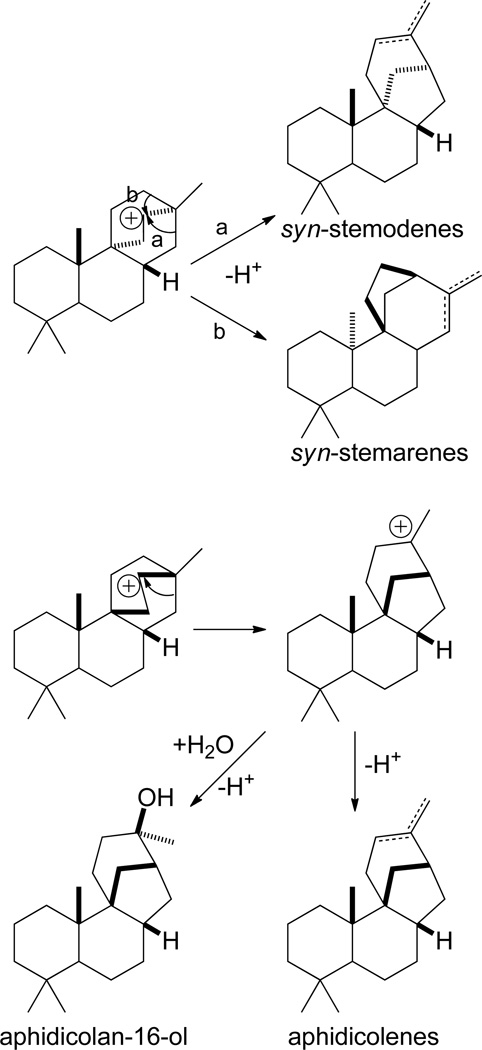
Examples of C-9,16 tetracyclization arise from both re- and si-face tricyclized syn-CPP following C-9,8 hydride shift within the corresponding (iso)pimaryl+ intermediates (Scheme 12). The secondary carbocationic intermediate formed by such tetracyclization seems to typically undergo ring rearrangement to a tertiary carbocationic intermediate, although its direct deprotonation also is plausible. In the case of the tetracycle derived from syn-pimar-15-en-9-yl+, formation of an aphidicol-16-yl+ intermediate is known. In particular, this intermediate can undergo direct deprotonation at either the neighboring methyl or endo-methylene, to yield the corresponding double bond isomers, and a mutant syn-pimaradiene synthase has been shown to selectively produce aphidicol-15-ene.55 Alternatively, this intermediate can undergo addition of water prior to deprotonation, and such an aphidicolan-16-ol forming diterpene synthase has been identified.32 The tetracycle derived from syn-isopimar-15-en-9-yl+ is known to undergo ring rearrangement from either of the neighboring endo-methylenes, with that from C-12 leading to a stemar-13-yl+ with a 6-6-5-6 ring structure analogous to that found in aphidicolanes, while ring rearrangement from C-14 results in stemodan-16-yl+, and both are observed to be directly deprotonated on the neighboring methyl or endo-methylene to the corresponding double bond isomers. Class I diterpene synthases forming either syn-stemodene and/or syn-stemarenes, more or less selectively, have been identified.56, 57 Formation of a 6-6-6-5 ring structure analogous to that of syn-stemarene from the tetracycle derived from syn-pimar-15-en-9-yl+ also is possible. Finally, an equivalent series of transformations can be envisioned for cyclization of syn-ent-CPP, with enantiomeric labdane-related diterpenoids having been isolated from Calceolaria species.33
Scheme 12.
3.4 Pentacyclic: Trachylobanes and Helifulvanes
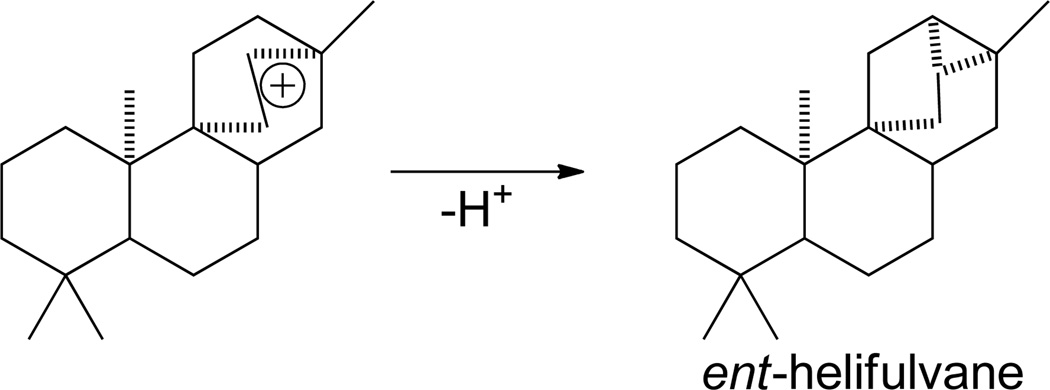
The formation of pentaycles occurs by deprotonation of delocalized rearrangements of the tetracycles, inevitably leaving a cyclopropyl ring.8, 9 The example of ent-trachylobane, derived from the delocalized ring rearrangement intermediate following C-8,16 tetracyclization, was described above (Scheme 11). Similarly, the pentacycle ent-helifuvane arises from C-9,16 tetracyclization and deprotonation of an analogous delocalized ring rearrangement intermediate (Scheme 13). However, while natural products with a helifulvane hydrocarbon backbone are known, in no such class I diterpene synthase activity been characterized.
Scheme 13.
3.5 Subsequent hydrocarbon backbone alterations: Gibberellins and Oryzalides
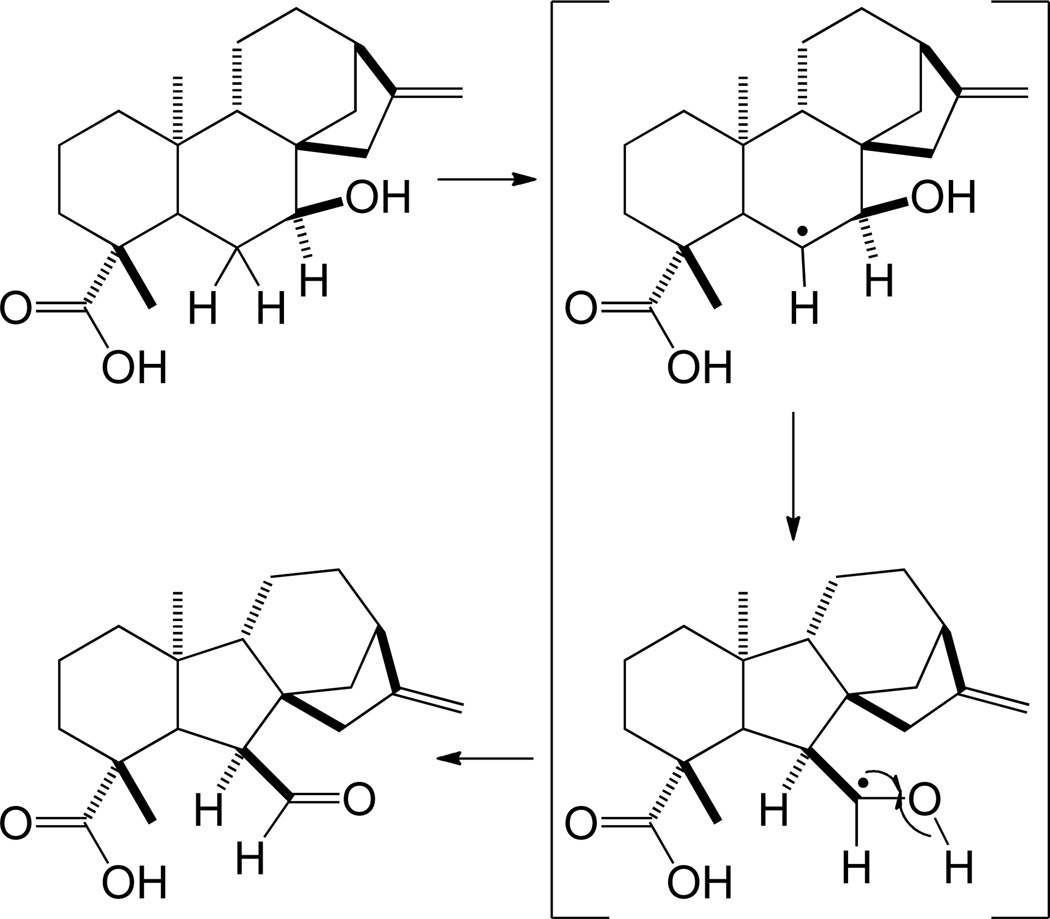
As noted above (Section 2.3), downstream transformations can alter the hydrocarbon ring structure formed by the initiating dual cyclization and/or rearrangement reactions. Both ring contraction and expansion are thought to occur. An example of ring expansion seems to arise from the presumably ent-atisirene derived aconitine diterpenoid crypto-alkaloid.58 More is known about the ring contraction reaction catalyzed in gibberellin metabolism.59 In particular, gibberellins are derived from ent-kaur-16-ene, with the ‘B’ ring of this 6-6-6-5 tetracycle contracted to a five membered ring to form the characteristic 6-5-6-5 ring structure of the gibberellins. This occurs following initial oxidation to ent-kaur-16-en-19-oic acid, with ‘B’ ring contraction occurring during the subsequent oxidation of the C-7 endo-methylene to a carboxylic acid, with both sets of oxidative reactions catalyzed by a cytochrome P450 mono-oxygenase, ent-kaurene oxidase and ent-kaurenoic acid oxidase, respectively. Accordingly, ring contraction is thought to be mediated by formation of a radical intermediate at C-6, with a 1,2 radical shift leading to extrusion of the previously formed C-7β-ol, and resulting in an aldehyde product (Scheme 14).9
Scheme 14.
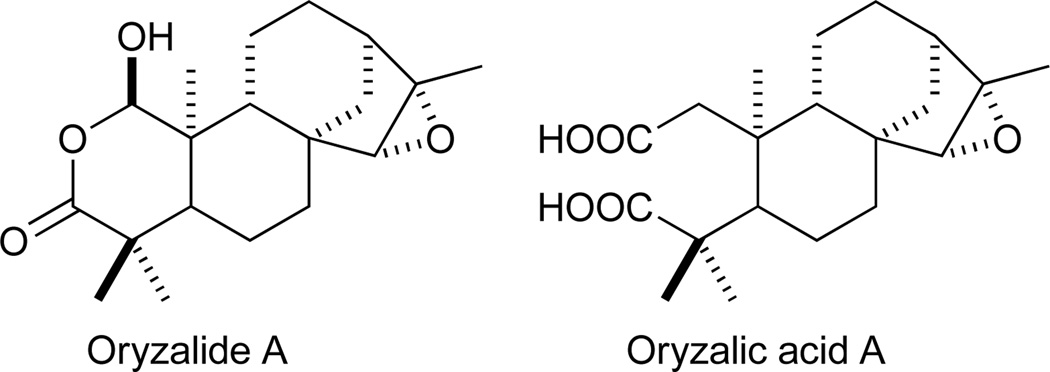
In addition to such direct ring rearrangements, it is possible for substitution within a carbocycle of the original hydrocarbon backbone to lead to heterocycle formation. An example of this can be seen in the antibacterial oryzalides found in rice.60, 61 These appear to be ent-isokaur-15-ene derived nor-diterpenoid natural products wherein C-2 of the ‘A’ ring has been substituted by an oxygen. Derivation of the oryzalides from ent-isokaurene can be readily discerned from the co-occurring oryzalic acids, wherein the ‘A’ ring has been fractured by the oxidation of both C-2&3 to carboxylic acids (Figure 2).61, 62
Figure 2.
4 Gibberellin phytohormone biosynthesis as a genetic reservoir
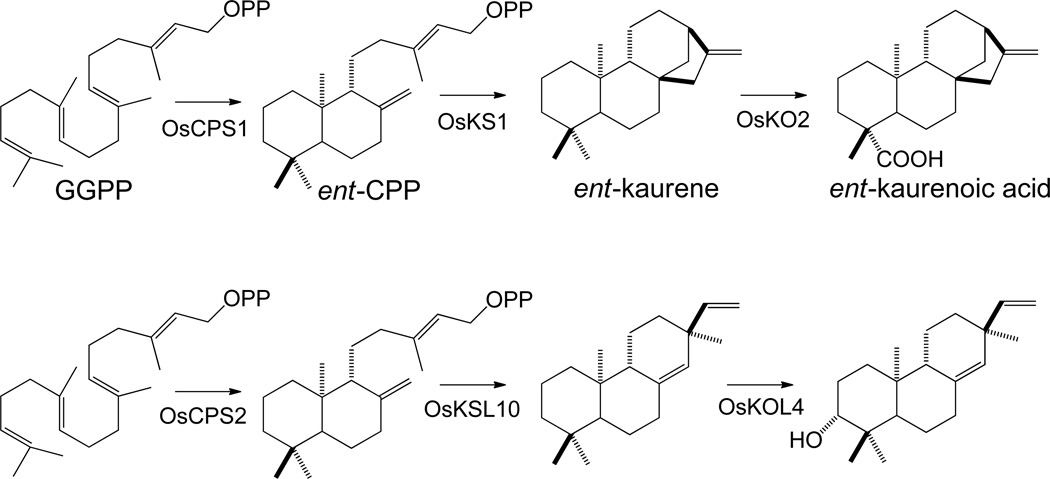
Among the labdane-related diterpenoids, the gibberellin phytohormones stand out for their critical role in normal plant growth and development. The requirement for gibberellin metabolism in all higher plants thus provides a genetic reservoir of biosynthetic genes from which other such natural products can and have been evolved. Not surprisingly then, the majority of labdane-related diterpenoids are found in plants. Gibberellin biosynthesis proceeds via cyclization of GGPP to ent-CPP and, hence, to the 6-6-6-5 tetracycle ent-kaurene, which is further oxidized to ent-kaurenoic acid prior to formation of the characteristic 6-5-6-5 gibberellin ring structure via oxidation of the C-7 endo-methylene to a carboxylic acid, as described above. Accordingly, the three early biosynthetic steps, i.e. those prior to formation of the gibberellin ring structure, provide direct access to ent-kaurene derived compounds (Scheme 15). Indeed, the more than 1,000 known such natural products are consistent with facile evolution of such alternative labdane-related diterpenoid biosynthesis.
Scheme 15.
Broader diversity also is readily accessible. For example, it has been shown that the ent-kaurene synthases found in all higher plants due to their necessary role in gibberellin biosynthesis can be converted to the production of tricyclic pimaradienes by a single amino acid change.53 In addition, the converse change can alter product outcome from a tricycle to tetracycle,55 and this single residue switch appears to be broadly applicable to class I diterpene synthase catalysis.63 Newly arisen labdane-related diterpenes might be recognized and oxygenated, increasing their polarity, solubility, and, hence, potential for exerting biological activity, either directly by the kaurene oxidase cytochrome P450 from gibberellin biosynthesis, or by other cytochromes P450 in the producing organisms. In this regard, it is notable that plants have vastly expanded numbers of cytochrome P450 mono-oxygenases in their genomes,64 providing a ready source of potential such downstream acting enzymes.
While the model plant Arabidopsis thaliana is thought to make only gibberellins, and not other labdane-related diterpenoids, rice (Oryza sativa) provides a model system that has long been recognized to make other such natural products.65 Recent work in rice is then providing insights into the evolution of alternative labdane-related diterpenoid biosynthesis from ancestral gibberellin metabolism. From comprehensive characterization of the class II and class I families of diterpene synthases, their homologous origins are readily evident.66 The rice class II diterpene cyclases have evolved such that even though there are two ent-CPP synthases, only one seems to play a role in gibberellin metabolism,67 with the other presumably required for alternative labdane-related diterpenoid biosynthesis,17 exhibiting closer homology to the syn-CPP synthase that must operate in such alternative metabolism.17, 18, 68 The rice class I diterpene synthases, while also all sharing homologous origins, have undergone more chemically divergent evolution, and produce a wide array of labdane-related diterpenes derived from the endogenous ent- or syn-CPP.42–46, 56, 57 Indeed, it was the functional genomics based investigations of the varied product outcome mediated by these rice ent-kaurene synthase-like paralogs that led to the insights into class I diterpene synthase plasticity discussed above, as well as resulting in identification of a number of novel such enzymes. Intriguingly, indirect evidence further suggests that some of the kaurene oxidase paralogs found in the rice genome might similarly act in such alternative labdane-related diterpenoid biosynthesis.69 Accordingly, work in rice has demonstrated the divergence of all three early acting enzymes from gibberellin metabolism to alternative labdane-related diterpenoid biosynthesis following gene duplication events, consistent with the derivative evolutionary scenario presented here (Scheme 15).
5 Driving diversity: Biological activity
While the discussion above emphasizes the facility with which alternative labdane-related diterpenoid biosynthesis can be derived from gibberellin metabolism, the fact that Arabidopsis lacks such expanded biosynthetic processes highlights the fact that this does not necessarily lead to such evolution, which must be driven by selective pressure. For example, the rice labdane-related diterpenoid natural products whose evolutionary origins are discussed above have putative roles as antimicrobial phytoalexins and/or neighboring plant growth suppressing allelochemicals.66 The selective pressure for retention of such metabolic capacity in rice is notably reflected in the fact that its genome contains two clusters of unrelated genes for labdane-related diterpenoid biosynthesis,16, 41, 68, 69 which is an unusual occurrence in plants that is associated with strong biological activity.70 Given the need for selective pressure to drive the evolution of alternative labdane-related diterpenoids, the large number of such natural products suggests that their underlying hydrocarbon skeletal structures might serve as privileged scaffolds from which biological activity is readily derived.
5.1 Repeated evolution of gibberellin biosynthesis
Perhaps the most striking example of the ability of selective pressure to drive natural products evolution arises from the intriguing observation that the potent effect of gibberellins on plant physiology have led to their production by not only plants, but plant associated microbes as well, both fungal and bacterial,71 with each of the relevant biological kingdoms appearing to have independently evolved such capacity.21 In all three cases, GGPP is cyclized to ent-kaurene, and it has been suggested that the relevant diterpene synthases may share some distant homologous origins.21 However, the remaining steps in fungal gibberellin biosynthesis are mediated by four cytochromes P450, which mediate early C-3 hydroxylation, along with a hydrocarbon desaturase.72 By contrast, the oxidative steps in plant gibberellin metabolism are catalyzed by a mixture of cytochromes P450 and 2-oxo-glutarate dependent dioxygenases, with C-3 hydroxylation occurring as the last step in production of the bioactive gibberellin phytohormones.73 Finally, while much less is known about bacterial GA production, it appears that only three cytochromes P450 and a short chain alcohol dehydrogenase are required.21 Notably, even in the case of the relevant cytochromes P450, across the various kingdoms these mono-oxygenases share no more than 15% amino acid sequence identity, while those involved in fungal gibberellin biosynthesis share more identity, indicating more homology between these consecutively acting enzymes than with their functional analogs from the other kingdoms. Thus, gibberellin biosynthesis in plants, fungi, and bacteria clearly represents an example of convergent metabolic evolution. Independent assembly of these complex multistep pathways is consistent with strong selective pressure for such biosynthesis arising from the potent effects of the bioactive gibberellins on plant physiology.
6 Conclusions
Based on their common biogenetic origins in the initiating dual cyclization and/or rearrangement reactions described above, a unifying labdane-related diterpenoid designation has been proposed here for the corresponding super-family of natural products. Intriguingly, while diterpene synthases catalyzing some of the possible cyclization and/or rearrangement reactions have been identified, many remain unknown. Even for those already identified, the enzymatic structure-function relationships underlying catalysis of what are often complex cyclization and rearrangement reactions, but which nevertheless generally lead to specific product outcome, is largely unknown, providing a rich area for continued investigation, as does study of the downstream tailoring enzymes.
Acknowledgements
Support for work on various aspects of labdane-related diterpenoid biosynthesis in the author’s laboratory from the NIH (GM076324 and GM086281), NSF (MCB0919735), and USDA-NIFA-NRI (2008-35318-05027) is gratefully acknowledged.
Biography
Reuben J. Peters received his B.S. from U.C. San Diego in 1992, and a Ph.D. with David Agard from U.C. San Francisco in 1998. After working as a Postdoctoral Fellow of the Jane Coffin Childs Memorial Fund for Medical Research with Rodney Croteau (Washington State University), Peters joined the faculty at Iowa State University in 2002, where he is currently an associate professor. His research interests focus on elucidation of the enzymatic mechanisms and metabolic pathways underlying biosynthesis of the labdane-related diterpenoid natural products defined here, as well as characterization of their physiological functions and other, potentially useful biological activities.
References
- 1.Croteau R, Kutchan TM, Lewis NG. In: Biochemistry & Molecular Biology of Plants. Buchanan B, Gruissem W, Jones R, editors. Rockville, MD, USA: Am. Soc. Plant Biologists; 2000. pp. 1250–1318. [Google Scholar]
- 2.Buckingham J. Dictionary of Natural Products (on-line web edition) Chapman & Hall/CRC Press; 2002. [Google Scholar]
- 3.Ruzicka L, Eschenmoser A, Heusser H. Experentia. 1953;IX:357–367. [Google Scholar]
- 4.Prisic S, Xu J, Coates RM, Peters RJ. ChemBioChem. 2007;8:869–874. doi: 10.1002/cbic.200700045. [DOI] [PubMed] [Google Scholar]
- 5.Upper CD, West CA. J. Biol. Chem. 1967;242:3285–3292. [PubMed] [Google Scholar]
- 6.This protonation-initiated cyclization reaction has been referred to in various ways. Originally and most commonly as type-B, to distinguish this from the more common allylic diphosphate ionizing/lysing terpene cyclization and/or rearrangement reaction, which is then designated type-A. However, this conflicts with the kaurene synthase A and B nomenclature originally used to differentiate the corresponding enzymes involved in gibberellin biosynthesis, which were the first identified (i.e. kaurene synthase-A catalyzes a type-B reaction, while kaurene synthase-B catalyzes a type-A reaction). Thus, the alternative class I and II designation suggested by Wendt and Schulz (ref. 7) is used here. In addition, reflecting the uniform catalysis of cyclization reactions by the relevant class II enzymes, these are designated more specifically as cyclases rather than using the more generic synthase nomenclature.
- 7.Wendt KU, Schulz GE. Structure. 1998;6:127–133. doi: 10.1016/s0969-2126(98)00015-x. [DOI] [PubMed] [Google Scholar]
- 8.Dev S, Misra R. Diterpenoids. Boca Raton, FL: CRC Press; 1985. [Google Scholar]
- 9.MacMillan J, Beale MH. In: Isoprenoids Including Carotenoids and Steroids. Cane DE, editor. Oxford: Elsevier Science Ltd; 1999. pp. 217–243. [Google Scholar]
- 10.Sun T-P, Kamiya Y. Plant Cell. 1994;6:1509–1518. doi: 10.1105/tpc.6.10.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ait-Ali T, Swain SM, Reid JB, Sun T, Kamiya Y. Plant J. 1997;11:443–454. doi: 10.1046/j.1365-313x.1997.11030443.x. [DOI] [PubMed] [Google Scholar]
- 12.Smith MW, Yamaguchi S, Ait-Ali T, Kamiya Y. Plant Physiol. 1998;118:1411–1419. doi: 10.1104/pp.118.4.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rebers M, Kaneta T, Kawaide H, Yamaguchi S, Yang YY, Imai R, Sekimoto H, Kamiya Y. Plant J. 1999;17:241–250. doi: 10.1046/j.1365-313x.1999.00366.x. [DOI] [PubMed] [Google Scholar]
- 14.Kawasaki T, Kuzuyama T, Kuwamori Y, Matsuura N, Itoh N, Furihata K, Seto H, Dairi T. J. Antibiot. 2004;57:739–747. doi: 10.7164/antibiotics.57.739. [DOI] [PubMed] [Google Scholar]
- 15.Bensen RJ, Johal GS, Crane VC, Tossberg JT, Schnable PS, Meeley RB, Briggs SP. Plant Cell. 1995;7:75–84. doi: 10.1105/tpc.7.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris LJ, Saparno A, Johnston A, Prisic S, Xu M, Allard S, Kathiresan A, Ouellet T, Peters RJ. Plant Mol. Biol. 2005;59:881–894. doi: 10.1007/s11103-005-1674-8. [DOI] [PubMed] [Google Scholar]
- 17.Prisic S, Xu M, Wilderman PR, Peters RJ. Plant Physiol. 2004;136:4228–4236. doi: 10.1104/pp.104.050567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otomo K, Kenmoku H, Oikawa H, Konig WA, Toshima H, Mitsuhashi W, Yamane H, Sassa T, Toyomasu T. Plant J. 2004;39:886–893. doi: 10.1111/j.1365-313X.2004.02175.x. [DOI] [PubMed] [Google Scholar]
- 19.Kawaide H, Imai R, Sassa T, Kamiya Y. J. Biol. Chem. 1997;272:21706–21712. doi: 10.1074/jbc.272.35.21706. [DOI] [PubMed] [Google Scholar]
- 20.Tudzynski B, Kawaide H, Kamiya Y. Curr. Gent. 1998;34:234–240. doi: 10.1007/s002940050392. [DOI] [PubMed] [Google Scholar]
- 21.Morrone D, Chambers J, Lowry L, Kim G, Anterola A, Bender K, Peters RJ. FEBS Lett. 2009;583:475–480. doi: 10.1016/j.febslet.2008.12.052. [DOI] [PubMed] [Google Scholar]
- 22.Ikeda C, Hayashi Y, Itoh N, Seto H, Dairi T. J. Biochem. 2007;141:37–45. doi: 10.1093/jb/mvm004. [DOI] [PubMed] [Google Scholar]
- 23.Keeling CI, Dullat HK, Yuen M, Ralph SG, Jancsik S, Bohlmann J. Plant Physiol. 2010;152:1197–1208. doi: 10.1104/pp.109.151456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sawada Y, Katsumata T, Kitamura J, Kawaide H, Nakajima M, Asami T, Nakaminami K, Kurahashi T, Mitsuhashi W, Inoue Y, Toyomasu T. J Exp Bot. 2008;59:3383–3393. doi: 10.1093/jxb/ern192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richman AS, Gijzen M, Starratt AN, Yang Z, Brandle JE. Plant J. 1999;19:411–421. doi: 10.1046/j.1365-313x.1999.00531.x. [DOI] [PubMed] [Google Scholar]
- 26.Stofer Vogel B, Wildung MR, Vogel G, Croteau R. J. Biol. Chem. 1996;271:23262–23268. doi: 10.1074/jbc.271.38.23262. [DOI] [PubMed] [Google Scholar]
- 27.Schepmann HG, Pang J, Matsuda SP. Arch. Biochem. Biophys. 2001;392:263–269. doi: 10.1006/abbi.2001.2438. [DOI] [PubMed] [Google Scholar]
- 28.Martin DM, Faldt J, Bohlmann J. Plant Physiol. 2004;135:1908–1927. doi: 10.1104/pp.104.042028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ro D-K, Bohlmann J. Phytochemistry. 2006;67:1572–1578. doi: 10.1016/j.phytochem.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 30.Toyomasu T, Niida R, Kenmoku H, Kanno Y, Miura S, Nakano C, Shiono Y, Mitsuhashi W, Toshima H, Oikawa H, Hoshino T, Dairi T, Kato N, Sassa T. Biosci Biotechnol Biochem. 2008;72:1038–1047. doi: 10.1271/bbb.70790. [DOI] [PubMed] [Google Scholar]
- 31.Gao W, Hillwig ML, Huang L, Cui G, Wang X, Kong J, Yang B, Peters RJ. Org. Lett. 2009;11:5170–5173. doi: 10.1021/ol902051v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oikawa H, Toyomasu T, Toshima H, Ohashi S, Kawaide H, Kamiya Y, Ohtsuka M, Shinoda S, Mitsuhashi W, Sassa T. J. Am. Chem. Soc. 2001;123:5154–5155. doi: 10.1021/ja015747j. [DOI] [PubMed] [Google Scholar]
- 33.Garbarino JA, Fraga BM, Hernandez MG, Chamy MC, Piovano M. Pure Appl. Chem. 2001;73:579–582. [Google Scholar]
- 34.Shephard F. Bristol: University of Bristol; 2002. [Google Scholar]
- 35.Wendt KU, Schulz GE, Corey EJ, Liu DR. Angew. Chem. Int. Ed. 2000;39:2812–2833. [PubMed] [Google Scholar]
- 36.Dairi T, Hamano Y, Kuzuyama Y, Itoh N, Furihata K, Seto H. J. Bact. 2001;183:6085–6092. doi: 10.1128/JB.183.20.6085-6094.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakano C, Okamura T, Sato T, Dairi T, Hoshino T. ChemComm. 2005;2005:1016–1018. doi: 10.1039/b415346d. [DOI] [PubMed] [Google Scholar]
- 38.Arigoni D. Pure Appl. Chem. 1968;17:331–348. doi: 10.1351/pac196817030331. [DOI] [PubMed] [Google Scholar]
- 39.Christianson DW. Chem. Rev. 2006;106:3412–3442. doi: 10.1021/cr050286w. [DOI] [PubMed] [Google Scholar]
- 40.Morrone D, Hillwig ML, Mead ME, Lowry L, Fulton DB, Peters RJ. Chem. Biol. submitted. [Google Scholar]
- 41.Wise ML, Savage TJ, Katahira E, Croteau R. J. Biol. Chem. 1998;273:14891–14899. doi: 10.1074/jbc.273.24.14891. [DOI] [PubMed] [Google Scholar]
- 42.Wilderman PR, Xu M, Jin Y, Coates RM, Peters RJ. Plant Physiol. 2004;135:2098–2105. doi: 10.1104/pp.104.045971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Otomo K, Kanno Y, Motegi A, Kenmoku H, Yamane H, Mitsuhashi W, Oikawa H, Toshima H, Itoh H, Matsuoka M, Sassa T, Toyomasu T. Biosci. Biotechnol. Biochem. 2004;68:2001–2006. doi: 10.1271/bbb.68.2001. [DOI] [PubMed] [Google Scholar]
- 44.Kanno Y, Otomo K, Kenmoku H, Mitsuhashi W, Yamane H, Oikawa H, Toshima H, Matsuoka M, Sassa T, Toyomasu T. Biosci. Biotechnol. Biochem. 2006;70:1702–1710. doi: 10.1271/bbb.60044. [DOI] [PubMed] [Google Scholar]
- 45.Xu M, Wilderman PR, Morrone D, Xu J, Roy A, Margis-Pinheiro M, Upadhyaya N, Coates RM, Peters RJ. Phytochemistry. 2007;68:312–326. doi: 10.1016/j.phytochem.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 46.Cho E-M, Okada A, Kenmoku H, Otomo K, Toyomasu T, Mitsuhashi W, Sassa T, Yajima A, Yabuta G, Mori K, Oikawa H, Toshima H, Shibuya N, Nojiri H, Omori T, Nishiyama M, Yamane H. Plant J. 2004;37:1–8. doi: 10.1046/j.1365-313x.2003.01926.x. [DOI] [PubMed] [Google Scholar]
- 47.Ravn MM, Coates RM, Jetter R, Croteau R. Chem. Commun. 1998;1998:21–22. [Google Scholar]
- 48.Peters RJ, Flory JE, Jetter R, Ravn MM, Lee H-J, Coates RM, Croteau RB. Biochemistry. 2000;39:15592–15602. doi: 10.1021/bi001997l. [DOI] [PubMed] [Google Scholar]
- 49.Yamaguchi S, Saito T, Abe H, Yamane H, Murofushi N, Kamiya Y. Plant J. 1996;10:101–111. doi: 10.1046/j.1365-313x.1996.10020203.x. [DOI] [PubMed] [Google Scholar]
- 50.Yamaguchi S, Sun T, Kawaide H, Kamiya Y. Plant Physiol. 1998;116:1271–1278. doi: 10.1104/pp.116.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Toyomasu T, Kawaide H, Ishizaki A, Shinoda S, Otsuka M, Mitsuhashi W, Sassa T. Biosci. Biotechnol. Biochem. 2000;64:660–664. doi: 10.1271/bbb.64.660. [DOI] [PubMed] [Google Scholar]
- 52.Hayashi K, Kawaide H, Notomi M, Sakigi Y, Matsuo A, Nozaki H. FEBS Lett. 2006;580:6175–6181. doi: 10.1016/j.febslet.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 53.Xu M, Wilderman PR, Peters RJ. Proc. Natl. Acad. Sci. U.S.A. 2007;104:7397–7401. doi: 10.1073/pnas.0611454104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robinson DR, West CA. Biochemistry. 1970;9:80–89. doi: 10.1021/bi00803a011. [DOI] [PubMed] [Google Scholar]
- 55.Morrone D, Xu M, Fulton DB, Determan MK, Peters RJ. J. Am. Chem. Soc. 2008;130:5400–5401. doi: 10.1021/ja710524w. [DOI] [PubMed] [Google Scholar]
- 56.Nemoto T, Cho E-M, Okada A, Okada K, Otomo K, Kanno Y, Toyomasu T, Mitsuhashi W, Sassa T, Minami E, Shibuya N, Nishiyama M, Nojiri H, Yamane H. FEBS Lett. 2004;571:182–186. doi: 10.1016/j.febslet.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 57.Morrone D, Jin Y, Xu M, Choi S-Y, Coates RM, Peters RJ. Arch. Biochem. Biophys. 2006;448:133–140. doi: 10.1016/j.abb.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 58.Wang F-P, Liang X-T. The Alkaloids. 2002;59:1–280. doi: 10.1016/s0099-9598(02)59008-8. [DOI] [PubMed] [Google Scholar]
- 59.Yamaguchi S. Annu Rev Plant Biol. 2008;59:225–251. doi: 10.1146/annurev.arplant.59.032607.092804. [DOI] [PubMed] [Google Scholar]
- 60.Watanabe M, Sakai Y, Teraoka T, Abe H, Kono Y, Uzawa J, Kobayashi K, Suzuki Y, Sakurai A. Agric. Biol. Chem. 1990;54:1103–1105. [Google Scholar]
- 61.Kono Y, Uzawa J, Kobayashi K, Suzuki Y, Uramoto M, Sakurai A, Watanabe M, Teraoka T, Hosokawa D. Agric. Biol. Chem. 1991;55:803–811. [Google Scholar]
- 62.Watanabe M, Kono Y, Uzawa J, Teraoka T, Hosokawa D, Suzuki Y, Sakurai A, Teraguchi M. Biosci Biotechnol Biochem. 1992;56:113–117. [Google Scholar]
- 63.Wilderman PR, Peters RJ. J. Am. Chem. Soc. 2007;129:15736–15737. doi: 10.1021/ja074977g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nelson DR, Schuler MA, Paquette SM, Werck-Reichhart D, Bak S. Plant Physiol. 2004;135(2):756–772. doi: 10.1104/pp.104.039826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kato T, Kabuto C, Sasaki N, Tsunagawa M, Aizawa H, Fujita K, Kato Y, Kitahara Y, Takahashi N. Tetrahedron Lett. 1973;14:3861–3864. [Google Scholar]
- 66.Peters RJ. Phytochemistry. 2006;67:2307–2317. doi: 10.1016/j.phytochem.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 67.Sakamoto T, Miura K, Itoh H, Tatsumi T, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Agrawal GK, Takeda S, Abe K, Miyao A, Hirochika H, Kitano H, Ashikari M, Matsuoka M. Plant Physiol. 2004;134:1642–1653. doi: 10.1104/pp.103.033696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu M, Hillwig ML, Prisic S, Coates RM, Peters RJ. Plant J. 2004;39:309–318. doi: 10.1111/j.1365-313X.2004.02137.x. [DOI] [PubMed] [Google Scholar]
- 69.Itoh H, Tatsumi T, Sakamoto T, Otomo K, Toyomasu T, Kitano H, Ashikari M, Ichihara S, Matsuoka M. Plant Mol. Biol. 2004;54:533–547. doi: 10.1023/B:PLAN.0000038261.21060.47. [DOI] [PubMed] [Google Scholar]
- 70.Osbourn AE, Field B. Cell. Mol. Life Sci. 2009;66:3755–3775. doi: 10.1007/s00018-009-0114-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fischbach MA, Clardy J. Nat. Biol. Chem. 2007;3:353–355. doi: 10.1038/nchembio0707-353. [DOI] [PubMed] [Google Scholar]
- 72.Tudzynski B. Appl. Microbiol. Biotechnol. 2005;66:597–611. doi: 10.1007/s00253-004-1805-1. [DOI] [PubMed] [Google Scholar]
- 73.Hedden P, Phillips AL, Rojas MC, Carrera E, Tudzynski B. J. Plant Growth Regul. 2001;20:319–331. doi: 10.1007/s003440010037. [DOI] [PubMed] [Google Scholar]