Abstract
The C-terminal —COOH of prenylated proteins is methylated to —COOCH3. The —COOCH3 ester forms are hydrolyzed by prenylated methylated protein methyl esterase (PMPMEase) to the original acid forms. This is the only reversible step of the prenylation pathway. PMPMEase has not been purified and identified and is therefore understudied. Using a prenylated-l-cysteine methyl ester as substrate, PMPMEase was purified to apparent homogeneity from porcine liver supernatant. SDS-PAGE analysis revealed an apparent mass of 57 kDa. Proteomics analyses identified 17 peptides (242 amino acids). A Mascot database search revealed these as portions of the Sus scrofa carboxylesterase, a 62-kDa serine hydrolase with the C-terminal HAEL endoplasmic reticulum-retention signal. It is at least 71% identical to such mammalian carboxylesterases as human carboxylesterase 1 with affinities toward hydrophobic substrates and known to activate prodrugs, metabolize active drugs, as well as detoxify various substances such as cocaine and food-derived esters. The purified enzyme hydrolyzed benzoyl-Gly-farnesyl-l-cysteine methyl ester and hydrocinamoyl farnesyl-l-cysteine methyl ester with Michaelis–Menten constant (Km) values of 33 ± 4 and 25 ± 4 µM and Vmax values of 4.51 ± 0.28 and 6.80 ± 0.51 nmol/min/mg of protein, respectively. It was inhibited by organophosphates, chloromethyl ketones, ebelactone A and B, and phenylmethylsulfonyl fluoride.
Keywords: Carboxymethyl Esterase, Prenylation, Organophosphates, Prenylated Proteins, Hydrolase, Ebelactone
INTRODUCTION
The estimated 2% of eukaryotic proteins that are prenylated [1] undergo S-adenosyl-l-methionine (SAM)-dependent methylation to form carboxylmethyl esters in a reaction catalyzed by prenylated protein methyl transferase (PPMTase). This reaction is the only reversible step in the prenylation pathway as the prenylated protein carboxylmethyl esters are then the substrates of the prenylation-dependent esterase, prenylated methylated protein methyl esterase (PMPMEase), which hydrolyzes the ester to form the original unmethylated proteins [2,3]. These reactions are significant with respect to the conformations of the prenylated proteins since the reversibility of the process entails the constant removal and reintroduction of a negative charge in the close vicinity of the isoprenyl moiety. The dynamic equilibrium may be critical to the biochemical activity of the proteins since the interaction of the prenylated proteins with other macromolecules may be significantly affected by the methylation state of the adjacent −COOH group. This is perhaps more substantial based on the fact that the isoprenyl group of proteins such as the G-protein γ subunits are essential for their coupling of receptor-derived signals to such effector enzymes as the adenylyl cyclases and phospholipase Cβ2 [2,4,5]. Furthermore, an isoprenyl-binding site has been identified using solution NMR on Rho dissociation inhibitor [6]. Prenyl-l-cysteine (PC) analogs that mimic only the C-terminal prenylated cysteine residue of prenylated proteins exert physiological effects such as the inhibition of platelet aggregation [2] or SAM-induced Parkinson’s disease-like effects in rats through mechanisms that do not involve the inhibition of the prenylated protein methyltransferase [7,8]. It is conceivable, therefore, that the methylated and demethylated forms of prenylated proteins may be variously preferred for functional interactions by different protein targets, thus rendering PPMTase and PMPMEase very important moderators of prenylated protein function.
The C-terminal cysteine residue with the S-conjugated isoprenyl group is structurally specific for interaction with other protein targets. For example, unlike l-AFC, farnesol, farnesal, and farnesylic acid are ineffective inhibitors of basal or receptor-mediated binding of GTP-γ-S to receptors on HL-60 granulocyte membranes [9]. This functional specificity may be a product of coevolution that could have extended to include interactions of the prenylated proteins and PC analogs to PMPMEase. Rat liver supernatant and brain membranes containing PMPMEase were previously found to hydrolyze prenylated substrates with Michaelis–Menten constants of 5 and 25 µM, respectively [10]. Using such prenylated cysteine ester substrates to screen chromatographic fractions, it would be possible to selectively enrich endogenous esterases that metabolize prenylated proteins.
Serine esterases have active site serine, histidine, and glutamate residues that form the catalytic triad involved in bond hydrolysis. During catalysis, the active site serine is temporarily acylated; the rapid hydrolysis of the acyl-enzyme intermediate ensures the continuous activity of the enzyme [11]. This mechanism is exploited by other compounds that, acting as pseudosubstrates for various hydrolytic enzymes, react with the serine residue resulting in very slow recovery rates with the net effect being that the covalently modified enzyme molecules are unable to further bind and hydrolyze substrates [11]. Organophosphorus pesticides (OPs) [11–13], lactones [14], sulfonyl fluorides [15], and chloromethylketones [16] inhibit various serine hydrolases by this mechanism. In previous studies, prenylated methylated protein methyl esterase from rat liver supernatant and brain membranes was inhibited by OPs [10]. Similarly, a prenylation specific esterase from bovine rod outer segment membranes was inhibited by ebelactone B[14].
In the current study, a carboxymethylated PC analog substrate assay was used to purify an esterase from porcine liver supernatant to apparent homogeneity. Tandem mass spectrometric analysis of the tryptic peptide digest and database Mascot search identified the enzyme as Sus scrofa carboxylesterase precursor. Its features that include the active site serine hydrolase catalytic triad of amino acids, the endoplasmic reticulum retention signal, and the large, flexible and hydrophobic active site of the highly homologous human form are consistent with the predicted properties of an OPs-sensitive, prenylation-dependent enzyme whose substrates are large proteins of diverse structures, respectively.
MATERIALS AND METHODS
Materials
Porcine liver was obtained from Bradley’s Country Stores (Tallahassee, FL) within 1 h of slaughter and frozen until required for enzyme preparation. Diethylaminoethyl (DEAE) sepharose, phenyl sepharose, chelating sepharose, cibacron blue sepharose, trans,trans-farnesol, benzil, and hydrocinnamoyl chloride were purchased from Sigma-Aldrich (St. Louis, MO). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels were from Fisher Scientific (Suwannee, GA).
Synthesis of Hydrocinnamoyl-farnesyl-l-cysteine Methyl Ester
Farnesyl-l-cysteine methyl ester (FCM) was synthesized as previously described [17]. The FCM (1 g) was dissolved in approximately 100 mL of acetone followed by the addition of triethylamine (TEA, 1 mL) and 1 g of hydrocinnamoyl chloride (Scheme 1). This was stirred for 2 h at room temperature. The solvent was removed under reduced pressure; the residue was dissolved in 150 mL of ethylacetate and washed three times with water. The ethylacetate layer was dried with anhydrous sodium sulfate and filtered, and the ethylacetate was removed under reduced pressure. The residue was purified by silica gel chromatography to obtain hydrocinnamoyl-farnesyl-l-cysteine methyl ester (HCFCM). The purified HCFCM was then analyzed by electron spray ionization mass spectrometry (ESI-MS).
SCHEME 1.
Synthesis of HCFCM and HCFC. HCFCM was synthesized from FCM and hydrocinnamoyl chloride as described in the methods section. This was then purified over silica gel, eluting with a gradient of hexane and acetone. Also shown is the hydrolysis of HCFCM to HCFC by PMPMEase.
Enzyme Preparation and Assay
The supernatant fraction was prepared from 100 g of porcine liver as previously described [10]. Assay for PMPMEase activity was conducted using 1 mM of either benzoyl-Gly-farnesyl-l-cysteine methyl ester (BzGFCM) or HCFCM substrate as previously described [10]. The supernatant was kept on ice and used for the purification of PMPMEase.
PMPMEase Purification
PMPMEase was purified from porcine liver supernatant using a succession of different purification media. Ice-cold porcine liver supernatant (8.8 g of protein in 1000 mL) was loaded onto a DEAE ion-exchange column (4.8 I.D. ×60 cm) that had previously been equilibrated with 20 mM Tris–HCl (pH 7.4) containing 0.1% Triton X-100 (buffer A) at a flow rate of 5 mL/min. This was followed by washing with buffer A until the UV absorbance at 280 nm subsided to the preloading values. The bound enzyme was then eluted from the column using a linear gradient from 0 to 1 M NaCl in buffer A over 2 h. Fractions (4.5 mL) were collected, and aliquots were assayed for enzyme activity using HCFCM as substrate. Fractions with enzymatic activity (fractions 121–310) were combined and subjected to further purification by hydrophobic interaction chromatography (HIC) on a column of phenyl sepharose. The pooled fractions from the DEAE column were supplemented with NaCl to a concentration of 1 M and then loaded at a flow rate of 1.5 mL/min onto the HIC column (2.5 cm I.D. × 30 cm) that had been preequilibrated with buffer A containing 1 M NaCl. After loading, the column was washed with the same buffer and the enzyme was eluted with a linear gradient from 1 to 0 M NaCl in the same buffer. Fractions (4.5 mL) were collected, and aliquots of the fractions were analyzed for enzyme activity. The fractions with enzymatic activity were combined and subjected to further purification on Ni2+-charged immobilized metal ion affinity chromatography (Ni-IMAC). The chelating resin was charged with Ni2+ ions and equilibrated with buffer A. The pooled enzyme containing fractions from the HIC step was applied to the Ni-IMAC column at a flow rate of 0.5 mL/min and washed with buffer A. Elution was conducted with an increasing gradient of histidine from 0 to 50 mM in the same buffer. Aliquots of the 4.5 mL fractions were assayed for PMPMEase activity. The combined active fractions were further purified on a Q-sepharose column (1 cm I.D. ×15 cm). The binding and elution was conducted with the same buffers as in the DEAE purification step except that the required volumes were significantly less. Fractions (4.5 mL) were collected, and aliquots were assayed for enzyme activity. Fractions with enzyme activity were combined and subjected to gel filtration chromatography on a Superdex 200 column (2 cm I.D. ×90 cm) that was preequilibrated and eluted with buffer A at a flow rate of 2.5 mL/min. Enzyme-containing fractions were further purified by Cu2+-charged immobilized metal ion affinity chromatography (Cu-IMAC), loading and washing in buffer A and eluting with a 0 to 50 mM gradient of histidine in buffer A. The fractions containing PMPMEase activity were combined and applied onto a Cibacron blue 3GA column (1 cm I.D. ×15 cm) that had been pre-equilibrated with buffer A. The flow-through contained virtually all of the PMPMEase enzymatic activity.
Protein Assays
The total protein concentration in various samples was measured using the bicinchoninic acid (BCA) protein assay reagent kit (Pierce, Rockford, IL) according to the supplier’s procedures. Bovine serum albumin was used as the standard.
SDS-PAGE Analysis
Aliquots (50 µL) of all protein samples were combined with 50 µL of SDS-PAGE sample buffer and boiled for 5 min. The samples were then separated on 4–12% gradient gels, and the protein bands were visualized by EZBlue coomassie staining (Sigma Chemical Co., St. Louis MO). SDS-PAGE molecular weight markers were used to calculate the molecular weight of the purified protein.
Protein Sequence Analysis
Purified PMPMEase from the last purification step (Cibacron Blue) was separated by SDS-PAGE (4%–12% gradient gels) electrophoresis followed by EZBlue coomassie staining. The protein bands were excised and submitted to Protana Analytical Services (Toronto, Ontario, Canada) for sequence analysis and protein identification. The protein bands were subjected to ingel trypsin digestion, peptide elution, and liquid chromatography coupled with tandem mass spectrometric (LC-MS/MS) analysis. The masses of the peptides were detected, and the peptides were further fragmented to obtain their individual sequences. The obtained peptide masses and sequences were then used in Mascot database searches for protein identification.
Effect of Serine Hydrolase Inhibitors on PMPMEase Activity
Representative compounds from various classes of chemicals known to inactivate serine hydrolases were tested against porcine liver PMPMEase. These included ebelactones, carbamates, chloromethylketones, phenylmethylsulfonyl fluoride (PMSF), and organophosphorus compounds. The compounds (in 5µL of dimethylformamide, 1 and 0.1 mM) were added to the enzyme (in 90 µL of 100 mM Tris–HCl buffer, pH 7.4) and preincubated for 20 min prior to the addition of the HCFCM substrate to a concentration of 1 mM. This was followed by further incubation and analysis as described in the “enzyme preparation and assay” section.
RESULTS
Synthesis of Hydrocinnamoyl-farnesyl-l-cysteine Methyl Ester
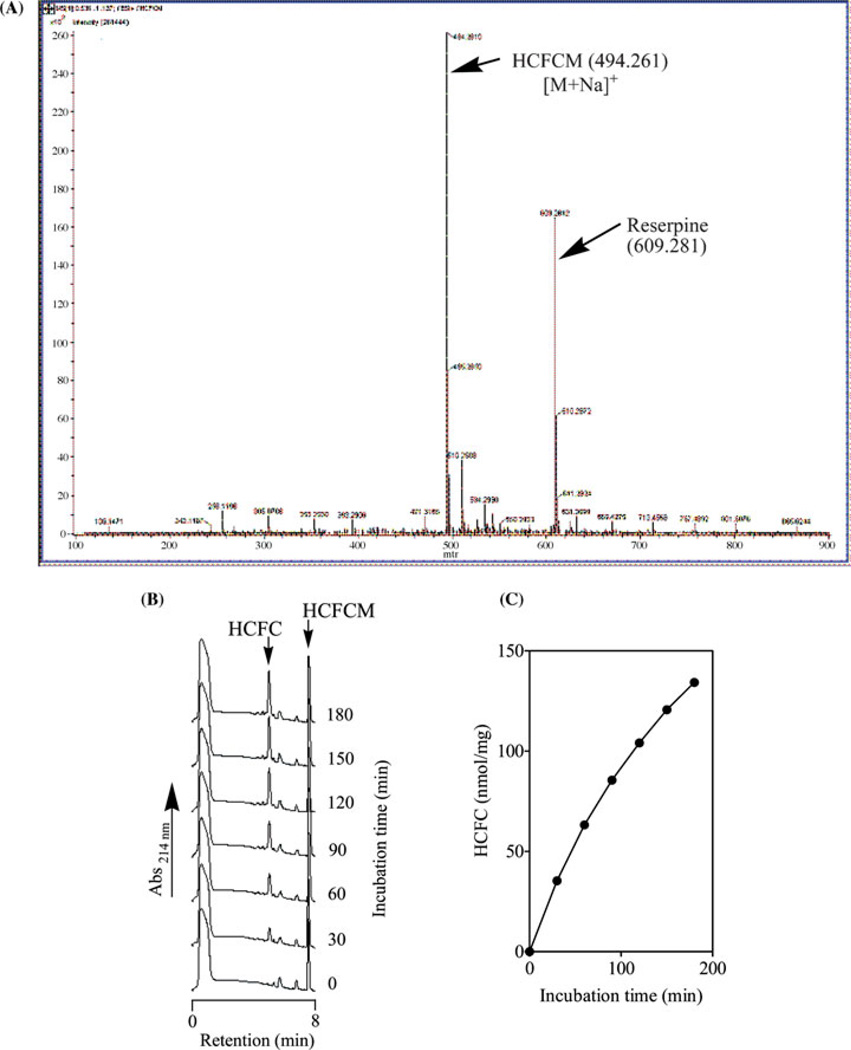
The reaction of FCM with hydrocinnamoyl chloride resulted in hydrocinamyl-S-farnesyl-l-cysteine methyl ester (HCFCM) as a waxy yellowish solid. When this was analyzed by ESI-MS, a single peak with a molecular weight of 494.26, corresponding to the sodium ion adduct of HCFCM, was observed (Figure 1A). When HCFCM (1 mM) was incubated with porcine liver supernatant, a time-dependent hydrolysis of the methyl ester to a single UV-absorbing product peak that coeluted with the free acid (hydrocinamylfarnesyl-l-cysteine, HCFC) was observed (Figures 1B and C). This indicates the hydrolysis of the ester bond of HCFCM to form HCFC. Hydrolysis was not observed in control incubations lacking enzyme. As expected given the S-farnesyl tail, HCFCM displayed comparable properties to BzGFCM with respect to the kinetics of hydrolysis by enzyme preparations.
FIGURE 1.
Mass spectrometry analysis of HCFCM (A). HCFCM (1 mM solution in methanol) was analyzed by electron spray ionization (ESI) mass spectrometry revealing a single distinct peak with an apparent relative molecular mass of 471.70. This corresponds to the calculated molecular mass of 471.70 (494.261 less the atomic mass for Na of 22.99). Reserpine was included in the analysis as an internal standard. Time-dependent demethylation of HCFCM to HCFC by pig liver supernatant PMPMEase (B and C). Porcine liver supernatant (0.4 mg of protein) was incubated with HCFCM (1 mM) in 100 mM Tris–HCl, pH 7.4 for the indicated times (right side of each chromatogram, panel A). Reactions were stopped with methanol and analyzed as indicated in the methods section. The results are the means (±SEM, n = 3. N.B. The error bars are obscured by the symbols).
Purification of Liver PMPMEase
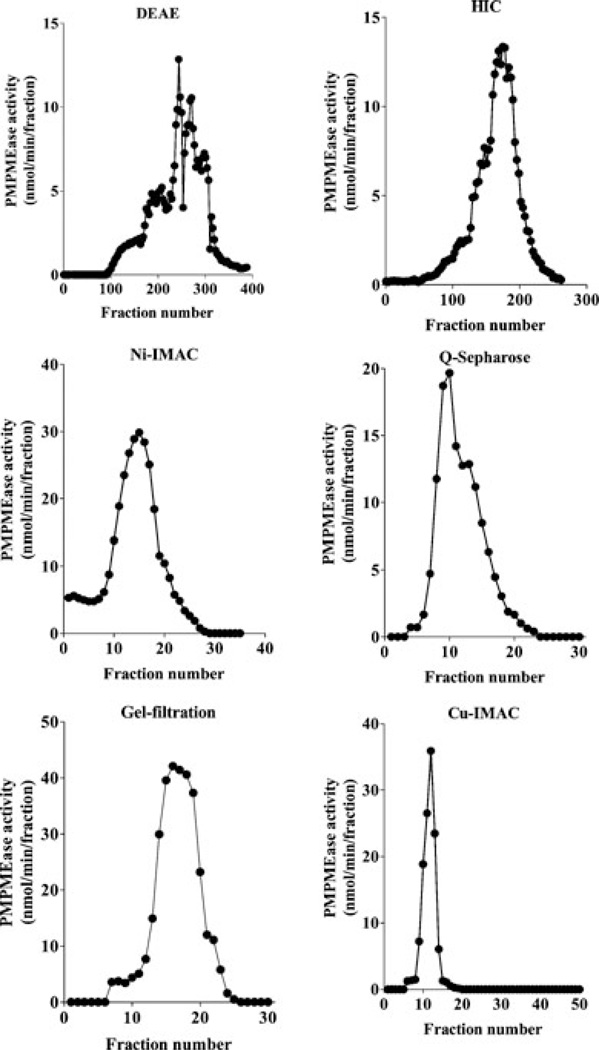
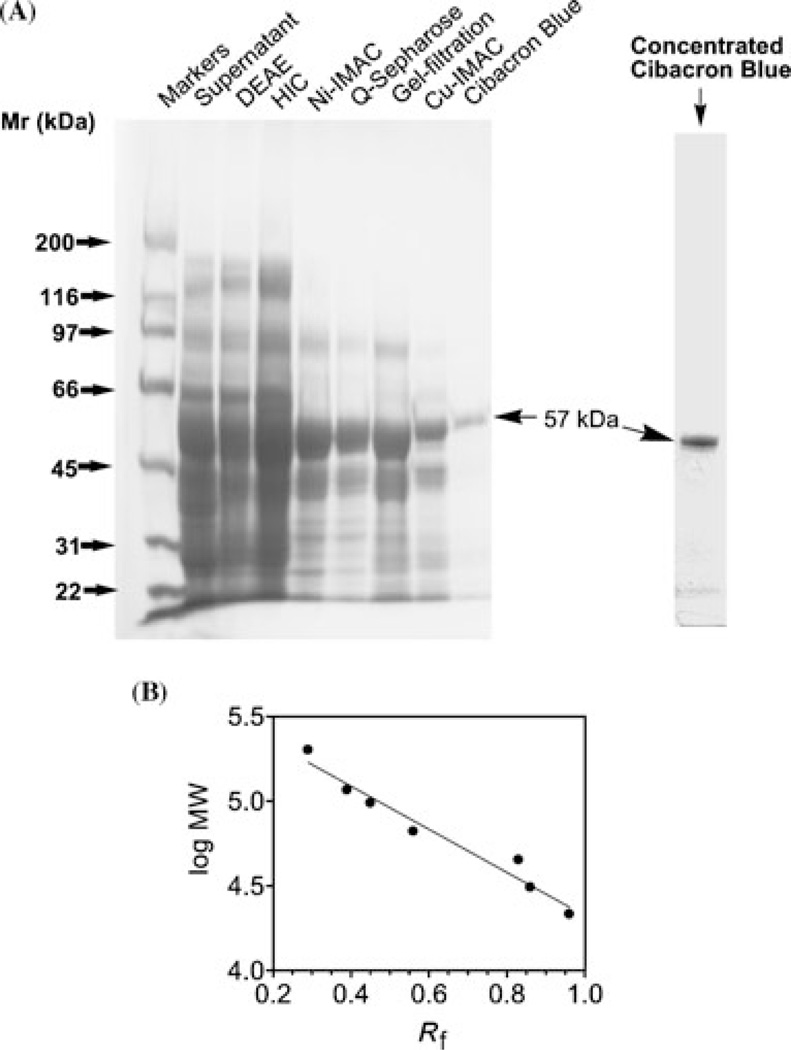
Various chromatographic media and conditions were used to sequentially purify porcine liver PMPMEase. As shown in Figure 2, PMPMEase bound to the different chromatographic media and was successfully eluted according to the procedures stated in the methods section. The enzyme did not bind to the Cibacron blue that was used in the last chromatographic step. The Cibacron blue did, however, appear to bind to some of the contaminating proteins as the enzyme flowed through. As shown in Table 1, the specific activity increased from 0.4 to 5.29 nmol of HCFCM hydrolyzed per min per mg of protein. This coincided with an over 13-fold enrichment of the PMPMEase activity. More than 80% of the enzyme activity was lost during the first purification step that involved ion exchange on DEAE sepharose (Table 1). SDS-PAGE protein analysis of samples from the different chromatographic steps revealed a successive elimination of contaminating protein bands that culminated in a single band of 57 kDa observed in the unbound fraction from the Cibacron blue step (Figure 3). This shows that although the enzyme did not bind to the Cibacron blue medium, it did indeed trap the remaining contaminating proteins from the Cu-IMAC column.
FIGURE 2.
Chromatographic purification of PMPMEase. All purification steps were conducted in 20 mM Tris–HCl, pH 7.4 containing 0.1% Triton X-100 as buffer A. All fractions were assayed for PMPMEase activity using HCFCM as the substrate. Porcine liver supernatant (8.8 g, 1000 mL) was applied to a DEAE-sepharose column and washed in buffer A. Bound proteins were eluted by an increasing linear gradient of 0 to 1 M NaCl in the buffer A. The PMPMEase-containing fractions from the DEAE step were applied to an HIC column in buffer A containing 1 M NaCl. Elution was achieved using a decreasing linear gradient of 1 to 0 M NaCl. The enzyme fractions from the HIC column were applied to the Ni-IMAC column. The enzyme was eluted with a linear gradient of 0 to 50 mM histidine in buffer A. The enzyme fractions were applied to a Q-sepharose column and eluted with a linear gradient of 0 to 1 M NaCl in buffer A. Fractions from the Q-sepharose column were further purified on a Superdex 200 gel-filtration column, eluting with 1 NaCl buffer A. The enzyme fractions were further purified on a Cu-IMAC column, eluting as in the case of Ni-IMAC. The resulting enzyme was then applied to a Cibacron blue column and eluted as described in the methods section.
TABLE 1.
Purification of Porcine Liver PMPMEase
| Chromatographic Step | Total Activity (nmol/min) |
Total Protein (mg) |
Specific Activity (nmol/min/mg) |
Purification Factor |
Yield (%) |
|---|---|---|---|---|---|
| Supernatant | 3485 | 8805 | 0.40 | 1.00 | |
| Diethylaminoethyl | 704 | 3113 | 0.23 | 0.57 | 20 |
| Hydrophobic interaction chromatography (HIC) | 667 | 484 | 1.38 | 3.48 | 19 |
| Ni2+-charged immobilized metal ion affinity chromatography | 374a | 265 | 1.41 | 3.57 | 11 |
| Q-sepharose | 444 | 220 | 2.02 | 5.09 | 13 |
| Gel-filtration | 221 | 75 | 2.96 | 7.47 | 6 |
| Cu2+-charged immobilized metal ion affinity chromatography (Cu-IMAC) | 150 | 60 | 2.49 | 6.28 | 4 |
| Cibacron blue | 127 | 5.29 | 13.23 | 3.6 |
Aliquots of the pooled enzyme containing fractions from each of thechromatographic steps were assayed for the total enzyme activity and total protein. The values obtained were then used to calculate the specificactivities, purification factors, and yields for the chromatographic steps.
Lower enzymatic activity than in the proceeding step possibly due to leached Ni2+ ions interfering with enzyme activity.
FIGURE 3.
SDS-PAGE analysis. (A) SDS-PAGE sample buffer (25 µL) was added to 25 µL aliquots of the combined fractions with PMPMEase activity from each of the chromatographic steps, boiled and separated on 4%–12% gradient SDS-PAGE gels followed by EZBlue coomassie staining. The Rf values for the molecular weight standards were determined and used to generate the calibration plot (panel B)usedtodetermine the molecular weightofthe single protein band from the Cibacron blue lane.
Protein Sequence Analysis
A single esterase of the serine hydrolase family was detected when SDS-PAGE gel slices of the purified PMPMEase were trypsinized and the peptide fragments were analyzed by LC-MS/MS followed by Mascot searches of sequence databases. As shown in Table 2, the masses and sequences of 17 tryptic-digest peptides were detected, which are identical to portions of the 62.3 kDa Sus scrofa carboxylesterase (pI of 5.69) or its precursor (pI of 5.62). The sequences of the 17 detected peptides amounted to 242 amino acids or 43% of the Sus scrofa carboxylesterase sequence (accession number NP-999411). Three peptides (FWANFAR, LGIWGFFSTGDEHSR and IPLQFSED-CLYLNIYTPADLTK) matched the Mus musculus esterase 22, an enzyme with similar sequence, molecular weight and pI characteristics as the Sus scrofa carboxylesterase. A protein identified as hypothetical protein from “Pongo pygmeus” matched the sequences of 10 of the peptides accounting for 22% of the entire 60-kDa protein. Both the Mus musculus esterase 22 and the Pongo pygmaeus hypothetical protein appear to be house mouse and orangutan species variants of the porcine carboxylesterase. Owing to the high sensitivity of LC-MS/MS analysis, other peptide sequences were detected that match 14 other proteins in sequence databases. Of these, none is an esterase, the only other detected hydrolase being aminoacylase 1 that hydrolyses acetyl groups from N-acetylated proteins [18].
TABLE 2.
Peptides Detected by In-Gel Trypsin Digestion of the Purified Enzyme Followed by LC-MS/MS Analysis
| Peptides Detected | Calculated Molecular Mass | Observed (M/Z) |
|---|---|---|
| FWANFAR | 910.44 | 456.60 |
| TATSLLWK | 918.52 | 460.33, 919.41 |
| QIAVLAGCK | 958.53 | 480.46 |
| TTTSAVFVHCLR | 1390.70 | 697.18 |
| QKSEDELLDLTLK | 1530.81 | 766.70 |
| FAPPQPAEPWSFVK | 1599.81 | 801.08 |
| LGIWGFFSTGDEHSR | 1708.65 | 855.33 |
| LKGEEVAFWNDLLSK | 1747.91 | 875.11 |
| SYPIANIPEELTPVATDK | 1957.00 | 979.70 |
| TVIGDHGDEIFSVFGFPLLK | 2190.14 | 1096.01 |
| MPEEILAEKDFNTVPYIVGINK | 2519.30 | 841.25 |
| YLGGTDDPVK | 1063.52 | 1064.41, 533.51 |
| AISESGVALTAGLVR | 1442.81 | 722.96 |
| GEEVAFWNDLLSK | 1506.74 | 754.69 |
| ESHPFLPTVVDGVLLPK | 1847.02 | 924.73 |
| GDAPEEEVSLSK | 1259.59 | 630.93 |
| YVSLEGLAQPVAVFLGVPFAKPPLGSLR | 2924.65 | 976.37 |
When the Sus scrofa amino acid sequence was used to conduct a BLAST database search, several carboxylesterases from different mammalian species and tissues were identified that show a 71%–80% identity and 83%–89% similarity to the Sus scrofa carboxylesterase 3 (Table 3). These have been reported to catalyze such reactions as the hydrolysis of triacylglycerols, cholesterol ester, cocaine, retinyl ester, as well as acyltransferase reactions. Until they are tested with prenylated ester substrates, the possibility that these highly homologous enzymes may be species/tissue variants of the porcine liver PMPMEase that might have coevolved with the ubiquitous prenylated proteins will remain unclear.
TABLE 3.
Sequence Identity and Similarity of the Purified Enzyme to Other Esterases
| Accession Number | Identity (%) | Similarity (%) | Reference | |
|---|---|---|---|---|
| Sus scrofa CES3 | NP 999411 | 100 | 100 | [23] |
| Canis familiaris CESdD1 | BAB60696 | 80 | 89 | [23] |
| Homo sapiens CES hBr2 | BAB85656 | 79 | 88 | Hosokawa et al., unpublisheda |
| Felis catus CES1, mRNA | BAC75712 | 78 | 87 | Miyazaki et al., unpublisheda |
| Bos taurus retinyl ester hydrolase (BREH1) | AAR14316 | 78 | 87 | Wu et al., unpublisheda |
| Macaca fascicularis CE | BAA24523 | 77 | 86 | Sone et al., unpublisheda |
| Homo sapiens CES1 (monocyte/macrophage) | P23141 | 77 | 87 | [44] |
| Homo sapiens acyl CoA: cholesterol acyltransferase | P23141 | 76 | 86 | [45] |
| Homo sapiens cholesterol ester hydrolase | AAP20868 | 76 | 86 | [46] |
| Oryctololagus cuniculus liver carboxylesterase | AAC39258 | 75 | 86 | [47] |
| Rattus norvegicus liver esterase (ES-10) | P16303 | 75 | 85 | [48] |
| Mesocricetus auratus liver carboxylesterase precursor | BAA05913 | 75 | 84 | [49] |
| Mus musculus triacylglycerol hydrolase | AAK58067 | 75 | 85 | [50] |
| Homo sapiens CES hBr3, brain carboxylesterase | BAA84996 | 74 | 84 | [51] |
| Mus musculus CES1 | AAH26897 | 71 | 83 | [52] |
A BLAST search was conducted with the full amino acid sequence of the Sus scrofa carboxylesterase precursor protein. Several sequences were detected, all with sequence identity and similarity to the query sequenceof atleast 71% and 83%, respectively. The proteins shownin the table were chosen for their reported substrates, species and tissue source, and for their sequence comparisons to the Sus scrofa carboxylesterase.
Available in the database with the respective sequences.
Hydrolysis of Farnesyl-l-cysteine Methyl Esters by Purified PMPMEase
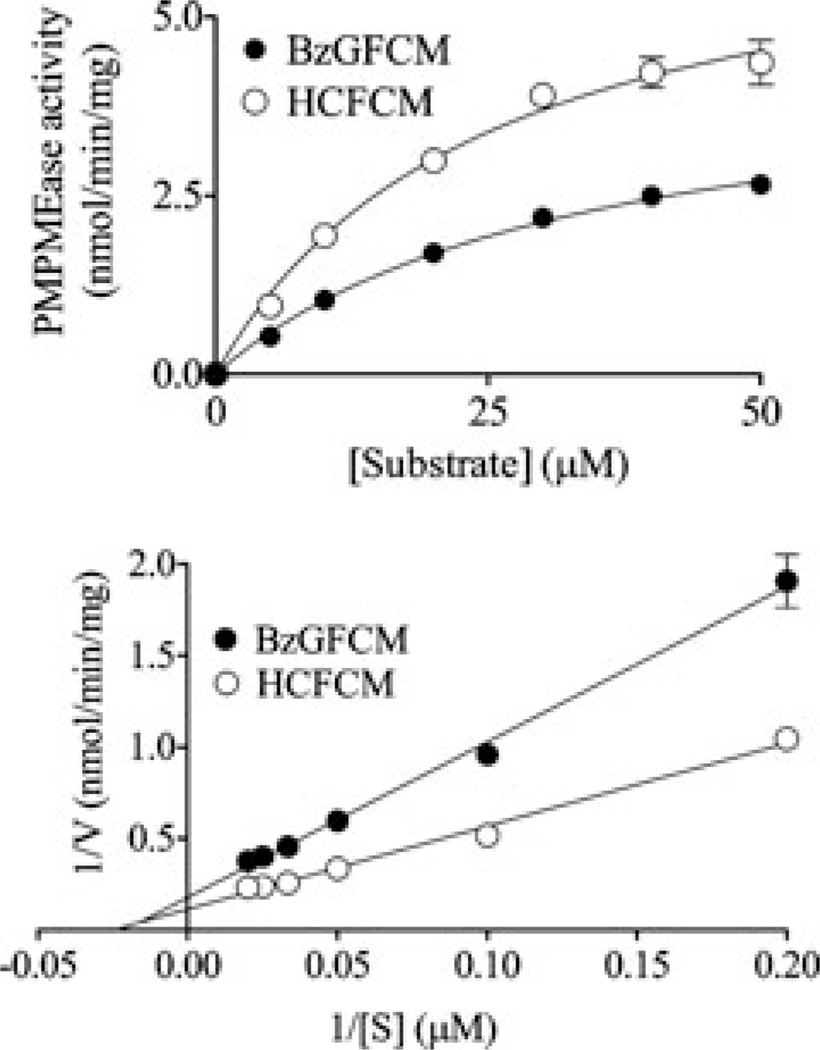
Purified PMPMEase hydrolyzed both BzGFCM and HCFCM in a concentration-dependent manner (Figure 4). Michaelis–Menten analysis of the substrate hydrolysis revealed Km values of 33 ± 4 and 25±4µM and Vmax values of 4.51 ±0.28 and 6.80 ±0.51 nmol/min/mg of protein for BzGFCM and HCFCM, respectively. The Km for BzGFCM using the purified porcine liver enzyme is about six-fold higher than that previously obtained using rat liver supernatant but comparable to that obtained with rat brain membrane bound enzyme [10].
FIGURE 4.
Michaelis–Menten kinetics (panel A) and double reciprocal plot (panel B) analysis of purified liver PMPMEase with prenyl-l-cysteine analog substrates. Purified liver PMPMEase (5.262 µg) was incubated with various concentrations of HCFCM (○) or BzGFCM (●) substrates as described in the methods section. The results are the means ±SEM of triplicate determinations.
Inhibition of Purified PMPMEase by Serine Hydrolase Inhibitors
The active site hydroxyl group of the catalytic serine residue of serine hydrolases is often the target of various compounds that covalently modify it resulting in the loss of enzymatic activity [13,19]. The compounds, thus acting as pseudosubstrates, react with the enzyme but unlike the normal carboxylester substrates, the pseudosubstrates are not converted to products. The longer duration of active site occupancy prevents access by the substrates thereby inhibiting the enzyme. Of the compounds tested against the purified enzyme, ebelactone B, paraoxon, and PMSF completely inhibited the purified enzyme at 1 mM concentrations (Table 4). Ebelactone A, l-I-4-tosylamino-2-phenylethyl chloromethyl ketone, and tosyl-l-phenylalanine chloromethyl ketone inhibited more than 80% of the enzyme at 1 mM concentrations. The other chloromethyl ketones inhibited about 45% of the activity at concentrations of 1 mM. The carbamate compounds did not inhibit the PMPMEase at the tested concentrations.
TABLE 4.
Inhibition of Purified PMPMEase by Serine-Hydrolase Inhibitors
| Residual Activity (% of Control, SEM, n = 3) |
||
|---|---|---|
| Compounds Tested | 1 mM | 0.1*/0.01 mM |
| Asulam | 91.7 ± 6.9 | 113 ± 6.3 |
| Benzil | 38.0 ± .43 | *69.4 ± .24 |
| Benzyl carbamate | 124 ± 5 | 117 ± 11 |
| Ebelactone A | 18.3 ± 0.3 | 123 ± 7 |
| Ebelactone B | 0 ± 0 | 56.4 ± 1.0 |
| l-Leu chloromethyl ketone (CMK) | 60.4 ± 1.9 | 93.3 ± 1.0 |
| l-I-4-Tosylamino-2-phenylethyl CMK | 18.0 ± 2.1 | 95.5 ± 3.0 |
| N-Carbobenzyloxy-l-Phe CMK | 57.0 ± 2.0 | 94.8 ± 1.3 |
| N-(Methoxysuccinyl)-l-Ala-l-Ala-l-Pro-l-Val CMK | 61.8 ± 2.0 | 105 ± 3.1 |
| N-α-Tosyl-l-Lys CMK | 98.6 ± 7.3 | 98.3 ± 1.9 |
| Tosyl-l-Phe CMK | 13.9 ± 1.1 | 96.5 ± 1.4 |
| (3S)-1-Chloro-3-tosylamido-7-amino-2-heptanone | 73.6 ± 1.2 | 99.1 ± 5.6 |
| Paraoxon | 0 ± 0 | 0 ± 0 |
| Phenylmethylsulfonylfluride | 0 ± 0 | 79.5 ± 1.8 |
Purified PMPMEase (10 µg) was incubated with 1, 0.1 (benzil only), and 0.01 mM concentrations of the indicated compounds for 20 min at 37°C. HCFCM (1 mM) was then added followed by further incubation and HPLC analysis.
DISCUSSION
Readily reversible secondary modifications such as kinase-catalyzed phosphorylation and phosphatase-catalyzed dephosphorylation reactions strongly influence both the conformations and the functions of various proteins. Such effects are only as impacting as the relative activity levels of the counteracting enzymes that metabolize them. In prenylated protein metabolism, two enzymes catalyze counteracting reactions in the final step of the pathway. An esterification reaction is counteracted by an esterase [2], thus ensuring a healthy physiological balance of the C-terminal uncharged methyl ester prenylated proteins and the charged carboxylate isoforms. The charge states of the carboxyl ends of the prenylated proteins may determine both their structures and the nature of the interactions with other proteins that are dependent on the polyisoprenyl moiety and vicinal groups. A polyisoprenyl-binding pocket was demonstrated on the Rho dissociation inhibitor [6] that strongly indicates the importance of the farnesyl or geranylgeranyl moiety and indeed of the prenylation pathway changes on protein–protein interactions. Furthermore, a strong influence on physiological well being was demonstrated with mice deficient in isoprenyl carboxylmethyl transferase (PPMTase) which, unable to methylate prenylated protein substrates, did not survive through midgestation [20]. Although this could, in some circumstances, be synonymous with an overactive PMPMEase that would ensure a lower proportion of prenylated and methylated proteins when compared to the unmethylated counterparts, the significance of PMPMEase on the physiological effects of prenylated proteins is yet to be determined. Its purification and identification is the very first step toward the understanding of its properties and how its levels of activity may influence cellular and physiological events other than the immediate hydrolysis of prenylated protein methyl esters.
The tandem mass spectrometry analysis and identification of the enzyme as a Sus scrofa carboxylesterase precursor is interesting in several respects. It shows high-sequence identity and even higher sequence similarity to various mammalian esterases that hydrolyze a wide range of mainly hydrophobic substrates and have molecular weights around 60 kDa [21–23]. These features are similar to those of the purified PMPMEase, which shows a molecular weight on SDS-PAGE of about 57 kDa, and both the rat liver [10] and porcine liver in this study show a high affinity for the hydrophobic PC analog ester substrates. The crystal structure of the human isoform, hCE1, reveals complexes of homotrimers at equilibrium with the homohexamers and the catalytic triad of Ser221–His468–Glu354 inside a deep hydrophobic pocket that has a small rigid subsite and a large flexible region [24,25]. Purification-induced dissociation of these multimeric complexes especially when high-salt concentrations were involved might have resulted in monomeric, enzymatically inactive conformations. This may thus explain the low yields of the purified enzyme activity since separate purifications of the enzyme with and without Triton X-100 resulted in similar losses in activity following the use of high-salt concentrations. The active site characteristics are believed to be adaptations for binding the structurally diverse set of substrates such as cocaine and heroin [24,26]. The enzyme has been described as being promiscuous due to its large and flexible active site that renders it the ability to hydrolyze a wide variety of substrates [24,27]. The toxic substances, narcotics, and pharmaceutical drugs and prodrugs that the carboxylesterases hydrolyze are not endogenous compounds. It is possible that the primary task of these esterases is to metabolize prenylated proteins which are indeed endogenous. The active site flexibility may be an adaptive feature that could have evolved to accommodate the structural diversity of the polypeptide portions of the prenylated protein substrates. These enzymes have been cloned/isolated from a wide range of tissues (see references in Table 4), a feature that is consistent with the ubiquitous nature of prenylated proteins. PMPMEase/Sus scrofa carboxylesterase, like the other highly homologous carboxylesterases, has the HXEL endoplasmic reticulum retention signal [28]. This retention signal has been reported in the sister enzyme, PPMTase/icmt, that catalyzes the forward prenylated protein methylation reaction [29]. This colocalization with the better characterized PPMTase/icmt further supports the notion that the esterase isolated in this study is indeed a prenylation pathway enzyme.
PMPMEase through its possible regulation of the functions of various types of prenylated proteins may exert profound effects on various intracellular events and consequently on animal physiology. Putative substrates include both heterotrimeric guanine nucleotide binding proteins (G-proteins), monomeric G-proteins, nuclear lamins, etc. [30]. These proteins mediate processes ranging from neurotransmitter signaling, cytoskeletal, and intracellular transportation functions, cell proliferation, differentiation, and apoptosis [30]. It could be inferred from these that aberrant levels of PMPMEase activity would be expressed through disease states such as cancers, neurodegenerative and neuropsychiatric disorders. In this regard, the identification of PMPMEase as a serine hydrolase is interesting with respect to and consistent with its susceptibility to OPs pesticides, benzil, PMSF, chloromethylketones, and ebelactones A and B. Exposure to OPs has been widely reported to cause delayed Parkinson’s disease (PD)-like dyskinesias in cases that survive the preceding rapid cholinergic poisoning [31–37]. The delayed effect may be due to a slow accumulation of the methylated prenylated proteins caused by the generalized inhibition of not only acetycholinesterase but also PMPMEase. In earlier studies in this laboratory, administration of SAM, the methyl donor coenzyme of PPMTase, caused the cardinal effects reminiscent of PD within minutes of injection [38]. These aberrant physiological effects, unlike human PD, were corrected within 2 h in the hitherto normal animals with an intact repertoire of enzymes to correct the artificial biochemical disturbance. These PD-like effects were later shown to be associated with the methylation of proteins of the heterotrimeric G-protein γ-subunit size range [7]. Furthermore, these PD-like effects were blocked by PC analogs, the synthetic mimics of the prenylated C-terminals of prenylated proteins [7,8]. A recent epidemiological study revealed that pesticides use increases the risk for developing PD by 70% [39]. It is worth noting that some OPs induce a delayed, chronic effect called organophosphate-induced delayed neuropathy (OPIDN) that is not observed in the afflicted individuals until 1–2 weeks after OPs poisoning and accompanied by the degeneration of long nerves from the distal ends toward the cell body [40]. In a recent study [41], researchers found that high levels of Rab1 expression protected dopaminergic neurons from α-synuclein-induced degeneration. Rab1 is a prenylated protein involved in intracellular vesicular trafficking [42]. If the delayed OPs effects on long nerves are due to their inhibition of esterases such as PMPMEase resulting in an impairment of the metabolism of prenylated proteins such as the Rab-CAAX subfamily of proteins that undergo methylation [43], it may help explain why long nerves that would rely most heavily on a well-organized intracellular transport system would be most susceptible and degenerate toward the source of genetic information which is the nucleus in the cell body.
Acknowledgments
Contract Grant Sponsor: NIH/NIGMS/MBRS/SCORE.
Contract Grant Number: GM 08111-35.
Contract Grant Sponsor: Pharmaceutical Research Center NIH/NCRR.
Contract Grant Number: G12 RR0 3020.
REFERENCES
- 1.Nalivaeva NN, Turner AJ. Post-translational modifications of proteins: Acetylcholinesterase as a model system. Proteomics. 2001;1:735–747. doi: 10.1002/1615-9861(200106)1:6<735::AID-PROT735>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 2.Ma YT, Shi YQ, Lim YH, McGrail SH, Ware JA, Rando RR. Mechanistic studies on human platelet isoprenylated protein methyltransferase: Farnesylcysteine analogs block platelet aggregation without inhibiting the methyltransferase. Biochemistry. 1994;33:5414–5420. doi: 10.1021/bi00184a009. [DOI] [PubMed] [Google Scholar]
- 3.Perez-Sala D, Tan EW, Canada FJ, Rando RR. Methylation and demethylation reactions of guanine nucleotide-binding proteins of retinal rod outer segments. Proc Natl Acad Sci USA. 1991;88:3043–3046. doi: 10.1073/pnas.88.8.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Myung CS, Yasuda H, Liu WW, Harden TK, Garrison JC. Role of isoprenoid lipids on the heterotrimeric G protein gamma subunit in determining effector activation. J Biol Chem. 1999;274:16595–16603. doi: 10.1074/jbc.274.23.16595. [DOI] [PubMed] [Google Scholar]
- 5.Dietrich A, Meister M, Brazil D, Camps M, Gierschik P. Stimulation of phospholipase C-beta 2 by recombinant guanine-nucleotide-binding protein beta gamma dimers produced in a baculovirus/insect cell expression system. Requirement of gamma-subunit isoprenylation for stimulation of phospholipase C. Eur J Biochem. 1994;219:171–178. doi: 10.1111/j.1432-1033.1994.tb19927.x. [DOI] [PubMed] [Google Scholar]
- 6.Gosser YQ, Nomanbhoy TK, Aghazadeh B, Manor D, Combs C, Cerione RA, Rosen MK. C-terminal binding domain of Rho GDP-dissociation inhibitor directs N-terminal inhibitory peptide to GTPases. Nature. 1997;387:814–819. doi: 10.1038/42961. [DOI] [PubMed] [Google Scholar]
- 7.Lamango NS, Charlton CG. Farnesyl-l-cysteine analogs block SAM-induced Parkinson’s disease-like symptoms in rats. Pharmacol Biochem Behav. 2000;66:841–849. doi: 10.1016/s0091-3057(00)00274-4. [DOI] [PubMed] [Google Scholar]
- 8.Lamango NS, Ayuk-Takem LT, Nesby R, Zhao WQ, Charlton CG. Inhibition mechanism of S-adenosyl-methionine-induced movement deficits by prenylcysteine analogs. Pharmacol Biochem Behav. 2003;76:433–442. doi: 10.1016/j.pbb.2003.08.017. [DOI] [PubMed] [Google Scholar]
- 9.Scheer A, Gierschik P. S-prenylated cysteine analogues inhibit receptor-mediated G protein activation in native human granulocyte and reconstituted bovine retinal rod outer segment membranes. Biochemistry. 1995;34:4952–4961. doi: 10.1021/bi00015a006. [DOI] [PubMed] [Google Scholar]
- 10.Lamango NS. Liver prenylated methylated protein methyl esterase is an organophosphate-sensitive enzyme. J Biochem Mol Toxicol. 2005;19:347–357. doi: 10.1002/jbt.20100. [DOI] [PubMed] [Google Scholar]
- 11.Glynn P. Neuropathy target esterase and phospholipid deacylation. Biochim Biophys Acta. 2005;1736:87–93. doi: 10.1016/j.bbalip.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 12.Costa LG. Current issues in organophosphate toxicology. Clin Chim Acta. 2006;366:1–13. doi: 10.1016/j.cca.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 13.Kwong TC. Organophosphate pesticides: Biochemistry and clinical toxicology. Ther Drug Monit. 2002;24:144–149. doi: 10.1097/00007691-200202000-00022. [DOI] [PubMed] [Google Scholar]
- 14.Tan EW, Rando RR. Identification of an isoprenylated cysteine methyl ester hydrolase activity in bovine rod outer segment membranes. Biochemistry. 1992;31:5572–5578. doi: 10.1021/bi00139a021. [DOI] [PubMed] [Google Scholar]
- 15.Kokotos G, Kotsovolou S, Constantinou-Kokotou V, Wu G, Olivecrona G. Inhibition of lipoprotein lipase by alkanesulfonyl fluorides. Bioorg Med Chem Lett. 2000;10:2803–2806. doi: 10.1016/s0960-894x(00)00566-7. [DOI] [PubMed] [Google Scholar]
- 16.Pasquato A, Pullikotil P, Asselin MC, Vacatello M, Paolillo L, Ghezzo F, Basso F, Di Bello C, Dettin M, Seidah NG. The proprotein convertase SKI-1/S1P. In vitro analysis of Lassa virus glycoprotein-derived substrates and ex vivo validation of irreversible peptide inhibitors J Biol Chem. 2006;281:23471–23481. doi: 10.1074/jbc.M513675200. [DOI] [PubMed] [Google Scholar]
- 17.Ma YT, Gilbert BA, Rando RR. Farnesylcysteine analogs to probe role of prenylated protein methyltransferase. Methods Enzymol. 1995;250:226–234. doi: 10.1016/0076-6879(95)50075-8. [DOI] [PubMed] [Google Scholar]
- 18.Uttamsingh V, Keller DA, Anders MW. Acylase I-catalyzed deacetylation of N-acetyl-l-cysteine and S-alkyl-N-acetyl-l-cysteines. Chem Res Toxicol. 1998;11:800–809. doi: 10.1021/tx980018b. [DOI] [PubMed] [Google Scholar]
- 19.Minton NA, Murray VS. A review of organophosphate poisoning. Med Toxicol Adverse Drug Exp. 1998;3:350–375. doi: 10.1007/BF03259890. [DOI] [PubMed] [Google Scholar]
- 20.Bergo MO, Leung GK, Ambroziak P, Otto JC, Casey PJ, Gomes AQ, Seabra MC, Young SG. Isoprenylcysteine carboxyl methyltransferase deficiency in mice. J Biol Chem. 2001;276:5841–5845. doi: 10.1074/jbc.C000831200. [DOI] [PubMed] [Google Scholar]
- 21.Sanghani SP, Davis WI, Dumaual NG, Mahrenholz A, Bosron WF. Identification of microsomal rat liver carboxylesterases and their activity with retinyl palmitate. Eur J Biochem. 2002;269:4387–4398. doi: 10.1046/j.1432-1033.2002.03121.x. [DOI] [PubMed] [Google Scholar]
- 22.Furihata T, Hosokawa M, Nakata F, Satoh T, Chiba K. Purification, molecular cloning, and functional expression of inducible liver acylcarnitine hydrolase in C57BL/6 mouse, belonging to the carboxylesterase multigene family. Arch Biochem Biophys. 2003;416:101–109. doi: 10.1016/s0003-9861(03)00286-8. [DOI] [PubMed] [Google Scholar]
- 23.David L, Guo XJ, Villard C, Moulin A, Puigserver A. Purification and molecular cloning of porcine intestinal glycerol-ester hydrolase-evidence for its identity with carboxylesterase. Eur J Biochem. 1998;257:142–148. doi: 10.1046/j.1432-1327.1998.2570142.x. [DOI] [PubMed] [Google Scholar]
- 24.Redinbo MR, Bencharit S, Potter PM. Human carboxylesterase 1: From drug metabolism to drug discovery. Biochem Soc Trans. 2003;31:620–624. doi: 10.1042/bst0310620. [DOI] [PubMed] [Google Scholar]
- 25.Fleming CD, Bencharit S, Edwards CC, Hyatt JL, Tsurkan L, Bai F, Fraga C, Morton CL, Howard-Williams EL, Potter PM, Redinbo MR. Structural insights into drug processing by human carboxylesterase 1: Tamoxifen, mevastatin, and inhibition by benzil. J Mol Biol. 2005;352:165–177. doi: 10.1016/j.jmb.2005.07.016. [DOI] [PubMed] [Google Scholar]
- 26.Bencharit S, Morton CL, Xue Y, Potter PM, Redinbo MR. Structural basis of heroin and cocaine metabolism by a promiscuous human drug-processing enzyme. Nat Struct Biol. 2003;10:349–356. doi: 10.1038/nsb919. [DOI] [PubMed] [Google Scholar]
- 27.Bencharit S, Morton CL, Hyatt JL, Kuhn P, Danks MK, Potter PM, Redinbo MR. Crystal structure of human carboxylesterase 1 complexed with the Alzheimer’s drug tacrine: From binding promiscuity to selective inhibition. Chem Biol. 2003;10:341–349. doi: 10.1016/s1074-5521(03)00071-1. [DOI] [PubMed] [Google Scholar]
- 28.Potter PM, Wolverton JS, Morton CL, Wierdl M, Danks MK. Cellular localization domains of a rabbit and a human carboxylesterase: Influence on irinotecan (CPT-11) metabolism by the rabbit enzyme. Cancer Res. 1998;58:3627–3632. [PubMed] [Google Scholar]
- 29.Romano JD, Schmidt WK, Michaelis S. The Saccha-romyces cerevisiae prenylcysteine carboxyl methyltransferase Ste14p is in the endoplasmic reticulum membrane. Mol Biol Cell. 1998;9:2231–2247. doi: 10.1091/mbc.9.8.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roskoski R., Jr Protein prenylation: A pivotal post-translational process. Biochem Biophys Res Commun. 2003;303:1–7. doi: 10.1016/s0006-291x(03)00323-1. [DOI] [PubMed] [Google Scholar]
- 31.Arima H, Sobue K, So M, Morishima T, Ando H, Katsuya H. Transient and reversible parkinsonism after acute organophosphate poisoning. J Toxicol Clin Toxicol. 2003;41:67–70. doi: 10.1081/clt-120018273. [DOI] [PubMed] [Google Scholar]
- 32.Kventsel I, Berkovitch M, Reiss A, Bulkowstein M, Kozer E. Scopolamine treatment for severe extra-pyramidal signs following organophosphate (chlorpyrifos) ingestion. Clin Toxicol (Phila) 2005;43:877–879. doi: 10.1080/15563650500357636. [DOI] [PubMed] [Google Scholar]
- 33.Shahar E, Bentur Y, Bar-Joseph G, Cahana A, Hershman E. Extrapyramidal parkinsonism complicating acute organophosphate insecticide poisoning. Pediatr Neurol. 2005;33:378–382. doi: 10.1016/j.pediatrneurol.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 34.Bhatt MH, Elias MA, Mankodi AK. Acute and reversible parkinsonism due to organophosphate pesticide intoxication: Five cases. Neurology. 1999;52:1467–1471. doi: 10.1212/wnl.52.7.1467. [DOI] [PubMed] [Google Scholar]
- 35.Muller-Vahl KR, Kolbe H, Dengler R. Transient severe parkinsonism after acute organophosphate poisoning. J Neurol Neurosurg Psychiatry. 1999;66:253–254. doi: 10.1136/jnnp.66.2.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davis KL, Yesavage JA, Berger PA. Single case study. Possible organophosphate-induced parkinsonism. J Nerv Ment Dis. 1978;166:222–225. doi: 10.1097/00005053-197803000-00010. [DOI] [PubMed] [Google Scholar]
- 37.Hsieh BH, Deng JF, Ger J, Tsai WJ. Acetylcholinesterase inhibition and the extrapyramidal syndrome: A review of the neurotoxicity of organophosphate. Neurotoxicology. 2001;22:423–427. doi: 10.1016/s0161-813x(01)00044-4. [DOI] [PubMed] [Google Scholar]
- 38.Lamango NS, Nesby RA, Charlton CG. Quantification of S-adenosylmethionine-induced tremors: A possible tremor model for Parkinson’s disease. Pharmacol Biochem Behav. 2000;65:523–529. doi: 10.1016/s0091-3057(99)00220-8. [DOI] [PubMed] [Google Scholar]
- 39.Ascherio A, Chen H, Weisskopf MG, O’Reilly E, McCullough ML, Calle EE, Schwarzschild MA, Thun MJ. Pesticide exposure and risk for Parkinson’s disease. Ann Neurol. 2006;60:197–203. doi: 10.1002/ana.20904. [DOI] [PubMed] [Google Scholar]
- 40.Glynn P. A mechanism for organophosphate-induced delayed neuropathy. Toxicol Lett. 2006;162:94–97. doi: 10.1016/j.toxlet.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 41.Cooper AA, Gitler AD, Cashikar A, Haynes CM, Hill KJ, Bhullar B, Liu K, Xu K, Strathearn KE, Liu F, Cao S, Caldwell KA, Caldwell GA, Marsischky G, Kolodner RD, Labaer J, Rochet JC, Bonini NM, Lindquist S. Alphasynuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson’s models. Science. 2006;313:324–328. doi: 10.1126/science.1129462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martinez O, Goud B. Rab proteins. Biochim Biophys Acta. 1998;1404:101–112. doi: 10.1016/s0167-4889(98)00050-0. [DOI] [PubMed] [Google Scholar]
- 43.Leung KF, Baron R, Ali BR, Magee AI, Seabra MC. Rab GTPases containing a CAAX motif are processed post-geranylgeranylation by proteolysis and methylation. J Biol Chem. 2007;282:1487–1497. doi: 10.1074/jbc.M605557200. [DOI] [PubMed] [Google Scholar]
- 44.Munger JS, Shi GP, Mark EA, Chin DT, Gerard C, Chapman HA. A serine esterase released by human alveolar macrophages is closely related to liver microsomal carboxylesterases. J Biol Chem. 1991;266:18832–18838. [PubMed] [Google Scholar]
- 45.Becker A, Bottcher A, Lackner KJ, Fehringer P, Notka F, Aslanidis C, Schmitz G. Purification, cloning, and expression of a human enzyme with acyl coenzyme A: Cholesterol acyltransferase activity, which is identical to liver carboxylesterase. Arterioscler Thromb. 1994;14:1346–1355. doi: 10.1161/01.atv.14.8.1346. [DOI] [PubMed] [Google Scholar]
- 46.Ghosh S. Cholesteryl ester hydrolase in human monocyte/macrophage: Cloning, sequencing, and expression of full-length cDNA. Physiol Genomics. 2000;2:1–8. doi: 10.1152/physiolgenomics.2000.2.1.1. [DOI] [PubMed] [Google Scholar]
- 47.Potter PM, Pawlik CA, Morton CL, Naeve CW, Danks MK. Isolation and partial characterization of a cDNA encoding a rabbit liver carboxylesterase that activates the prodrug irinotecan (CPT-11) Cancer Res. 1998;58:2646–2651. [PubMed] [Google Scholar]
- 48.Robbi M, Beaufay H, Octave JN. Nucleotide sequence of cDNA coding for rat liver pI 6.1 esterase (ES-10), a carboxylesterase located inthe lumen of the endoplasmic reticulum. Biochem J. 1990;269:451–458. doi: 10.1042/bj2690451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sone T, Isobe M, Takabatake E, Wang CY. Cloning and sequence analysis of a hamster liver cDNA encoding a novel putative carboxylesterase. Biochim Biophys Acta. 1994;1207:138–142. doi: 10.1016/0167-4838(94)90063-9. [DOI] [PubMed] [Google Scholar]
- 50.Dolinsky VW, Sipione S, Lehner R, Vance DE. The cloning and expression of a murine triacylglycerol hydrolase cDNA and the structure of its corresponding gene. Biochim Biophys Acta. 2001;1532:162–172. doi: 10.1016/s1388-1981(01)00133-0. [DOI] [PubMed] [Google Scholar]
- 51.Mori M, Hosokawa M, Ogasawara Y, Tsukada E, Chiba K. cDNA cloning, characterization and stable expression of novel human brain carboxylesterase. FEBS Lett. 1999;458:17–22. doi: 10.1016/s0014-5793(99)01111-4. [DOI] [PubMed] [Google Scholar]
- 52.Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, Wagner L, Shenmen CM, Schuler GD, Altschul SF, Zeeberg B, Buetow KH, Schaefer CF, Bhat NK, Hopkins RF, Jordan H, Moore T, Max SI, Wang J, Hsieh F, Diatchenko L, Marusina K, Farmer AA, Rubin GM, Hong L, Stapleton M, Soares MB, Bonaldo MF, Casavant TL, Scheetz TE, Brownstein MJ, Usdin TB, Toshiyuki S, Carninci P, Prange C, Raha SS, Loquellano NA, Peters GJ, Abramson RD, Mullahy SJ, Bosak SA, McEwan PJ, McKernan KJ, Malek JA, Gunaratne PH, Richards S, Worley KC, Hale S, Garcia AM, Gay LJ, Hulyk SW, Villalon DK, Muzny DM, Sodergren EJ, Lu X, Gibbs RA, Fahey J, Helton E, Ketteman M, Madan A, Rodrigues S, Sanchez A, Whiting M, Young AC, Shevchenko Y, Bouffard GG, Blakesley RW, Touchman JW, Green ED, Dickson MC, Rodriguez AC, Grimwood J, Schmutz J, Myers RM, Butterfield YS, Krzywinski MI, Skalska U, Smailus DE, Schnerch A, Schein JE, Jones SJ, Marra MA. Generation and initial analysis of more than 15000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci USA. 2002;99:16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]