Figure 3. The Open Conformer Releases LysRS From MSC.
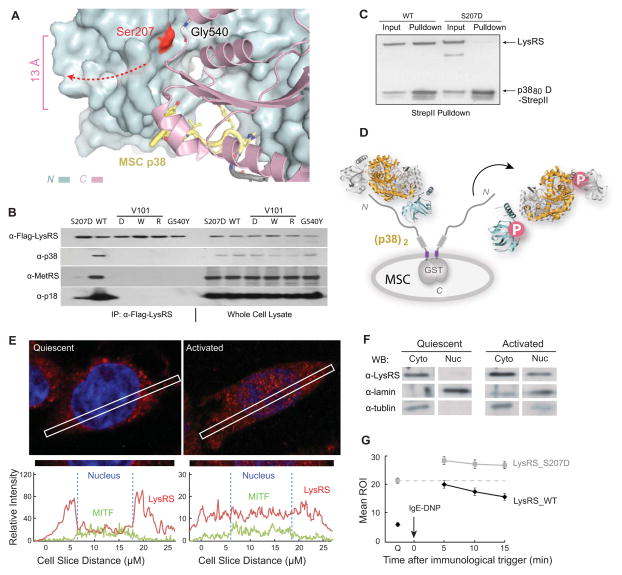
(A) View of the LysRS:p38/AIMP2 interface showing that the Ser207 phosphorylation site is at the inter-domain interface that forms the p38/AIMP2 binding groove. Ser207 is 13 Å away from the bound p38/AIMP2.
(B) Co-immunoprecipitation analyses showing that mutations of LysRS predicted to affect the p38/AIMP2 interaction (V101D, V101W, V101R), a mutation causing structural opening (G540Y) and the phospho-mimetic mutation (S207D), all dissociate LysRS from the endogenous MSC in HEK293T cells.
(C) Pull-down assay showing LysRSS207D loses the ability to bind a recombinant version of p38/AIMP2.
(D) Mechanism of Ser207 phosphorylation-directed release of LysRS from the MSC: The flipped-out anticodon-binding domain in LysRSpS207 opens the binding groove for p38/AIMP2 and prevents LysRS-p38/AIMP2 interaction.
(E) Confocal immunofluorescence microscopy showing the nuclear localization of the endogenous LysRS (red) and MITF (green) in quiescent RBL mast cells and following an IgE-DNP trigger. The blue signal is nuclear DAPI. The picture was taken by a confocal microscopy in Z-mode. The amount of LysRS and MITF signal in the cross section is quantified at the bottom.
(F) Nuclear localization of LysRS following RBL mast cell activation. Western blot analysis of LysRS in cytoplasmic versus nuclear fractions in quiescent mast cells versus in cells activated by the IgE-DNP trigger for 5 min.
(G) Confocal live imaging quantification of LysRS nuclear translocation. Means ROI (region of illuminated) of nuclear eGFP-LysRSwt versus eGFP-LysRSS207D at quiescent state (Q) and 5, 10, 15 min after immune activation. The means and standard errors of the means for 20 representative cells are shown (P<0.05).
See also Figure S4