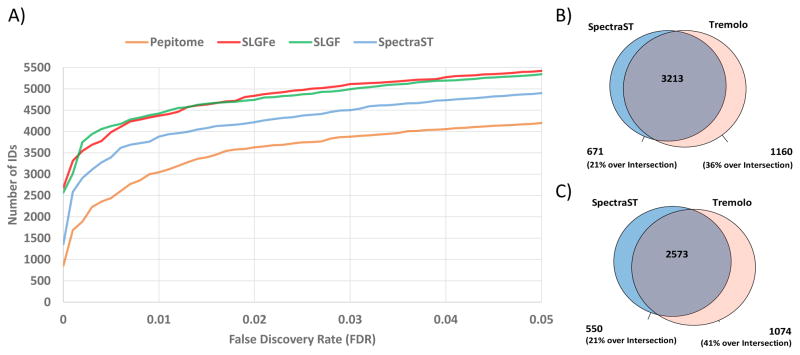
Figure 4.
Peptide spectral library search sensitivity and specificity comparison between Tremolo, SpectraST, and Pepitome. In (A) the performance of the scoring function SLGF is shown to be comparable to that of SLGFe, where one explicitly attempts to not consider mixture spectra (see textb for details). This contrasts to the performance of SpectraST in blue, and Pepitome in orange. (B) Number of spectra identified exclusively by SpectraST, Tremolo, and by both tools at 1% spectrum level FDR. On this Test dataset, SLGFe was also found to be more sensitive than SpectraST across the whole range of FDR thresholds (> 12% increase in IDs at 1% FDR). (C) Number of peptide IDs at 1% FDR by SpectraST and Tremolo. Tremolo identified 16% more peptides than SpectraST.