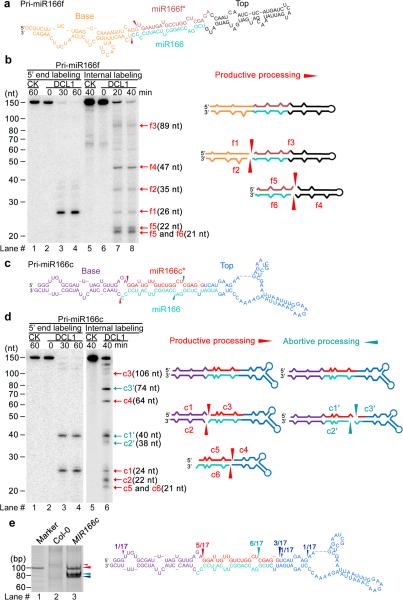
Figure 2.
Concomitant presence of productive and abortive processing of pri-miR166c but not of pri-miR166f. (a) Predicted secondary structure of in vitro transcribed pri-miR166f. Red arrows indicate the expected initial processed sites of pri-miR166f. (b) In vitro DCL1 reconstitution assay with the 5′ end or internally labeled pri-miR166f transcripts. Immunoprecipitates were prepared through two-step affinity purification using antibodies to Flag and then to Myc from N. bentha expressing 2Flag-4Myc-DCL1, 6Myc-HYL1, and SE-3HA complexes (DCL1) and prepared from mock-infiltrated plants (CK, referring to control check). RNAs recovered from the reaction mix were fractionated on 15% denaturing gels and detected by phosphor imaging. The positions of intact substrates, cleavage products, and reaction time are marked. Schematic illustration of the intermediate and final cleavage products (f1-f6) of pri-miR166f by DCL1 is shown on right. (c) Predicted secondary structure of in vitro transcribed pri-miR166c. Red and turquoise arrows show the expected initial productive processing and the unexpected abortive processing sites of pri-miR166c, respectively. (d) In vitro DCL1 reconstitution assays with 5′ end or internally labeled pri-miR166c transcripts. The assays were conducted as in Panel (b). Schematic illustration of the intermediate and final cleavage products (c1-6 and c1′-c3′) of pri-miR166c by DCL1 is shown on right. (e) Productive- and abortive-processing fragments were detected by 5′ RACE experiments in transgenic plants expressing 35S-MIR166c in vivo. The PCR products from the 5′ RACE experiments were recovered, sequenced, and mapped on the pri-miR166c and their counts are shown on right. Related uncropped images can be found Supplementary 9.