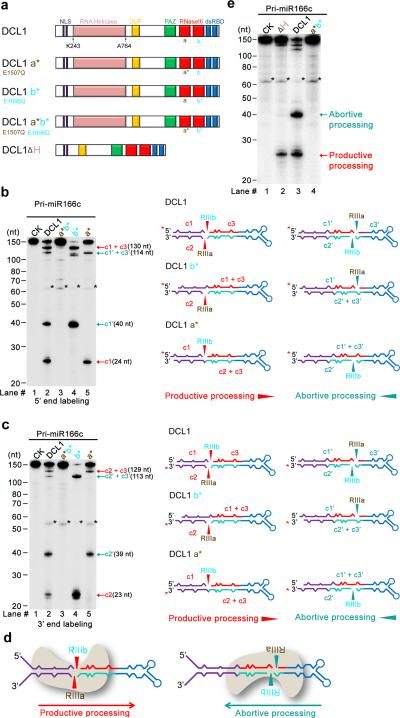
Figure 3.
DCL1 complexes process pri-miR166c bi-directionally: from lower stem to loop and from loop to lower stem. (a) Schematic illustration of DCL1 domains and DCL1 mutants. DCL1 a*, b*, a* b*, and DCL1 (ΔH) refer to RNaseIII a (E1507Q), b (E1696Q) point mutation, double mutations (E1507Q E1696Q), and a helicase-domain deletion mutant, respectively. (b) In vitro cleavage assays with the 5′ end labeled pri-miR166c transcripts by DCL1 and DCL1 mutants. Immunoprecipitation, cleavage assays, and RNA processing were performed as in Figure 2b. Black asterisks show the residual non-specific cleavage frequently present in the assay system. Schematic illustration of the cleavage products is shown on right. Red asterisks on transcripts indicate the 32P-labelling positions. Red and turquoise arrows show the productive and abortive processing sites, respectively. (c) In vitro cleavage assays with the 3′ end labeled pri-miR166c transcripts by DCL1 and DCL1 mutants. The cleavage assays and schematic illustration were essentially the same as in Panel (b). Note: the 3′ end labeling added extra “Cytosine” on the 3′ end of pri-miR166c and thus, the lengths of c2 and c2′ fragments in Figure 3c are 1-nt longer than those in Figure 2d. (d) Schematic for two orientations of DCL1 in processing of pri-miR166c and resultant constructive or destructive processing patterns. (e) In vitro cleavage assays with the 5′ end labeled pri-miR166c transcripts by DCL1(ΔH). The cleavage assays and schematic illustration were the same as in Panel (b). Related uncropped images can be found Supplementary 9.