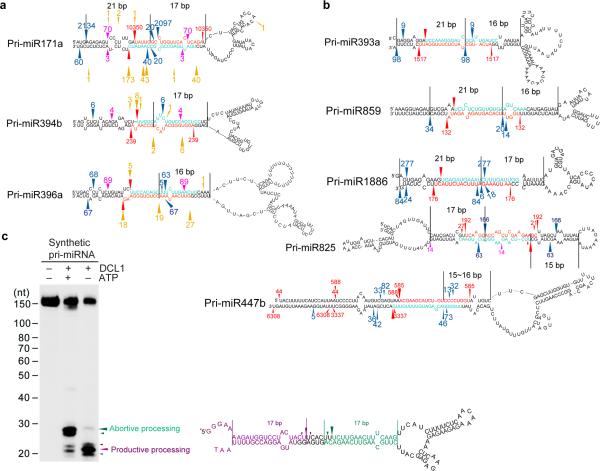
Figure 7.
Bi-directional processing of pri-miRNAs (BTLs) extends beyond pri-miR166 family in Arabidopsis. (a) Examples recovered from analyses of degradome and sRNA libraries. (b) Examples recovered only from analysis of sRNA libraries. For panels (a and b), miRNA and mRNA* sequences are marked in turquoise and red. Red arrows ( ) and numbers show productive processing sites and counts of miRNA* reads. Blue arrows (
) and numbers show productive processing sites and counts of miRNA* reads. Blue arrows ( ) and numbers show positions and counts of sRNAs resulting from apparent abortive processing events. Pink arrows (
) and numbers show positions and counts of sRNAs resulting from apparent abortive processing events. Pink arrows ( ) and numbers show positions and counts of sRNAs resulting from yet unappreciated processing events. Gold arrows (
) and numbers show positions and counts of sRNAs resulting from yet unappreciated processing events. Gold arrows ( ) and numbers show 5′ positions and counts of degradome reads22,41-43. The distance from abortive processing sites to terminal loops or between processing sites is shown between two black lines. Note: reads of miRNAs are not shown because they are not unique for individual pri-miRNA paralogs. Minor forms of miRNA/*s that are progressively away from predominant miRNA/* loci are not shown and can be referred to in Supplemental Tables 2 and 3. (c) In vitro DCL1 reconstitution assays with a synthetic TLBed pri-miRNA. Schematic illustration of the cleavage products by DCL1 is shown on right. Protein immunoprecipitation, cleavage assays and RNA processing were performed as described in Figure 2b. The positions of intact substrates, cleavage products, and RNA markers are shown. Red asterisks on transcripts indicate the 32P-labelling positions. Turquoise and purple arrows indicate abortive and productive processing sites, respectively.
) and numbers show 5′ positions and counts of degradome reads22,41-43. The distance from abortive processing sites to terminal loops or between processing sites is shown between two black lines. Note: reads of miRNAs are not shown because they are not unique for individual pri-miRNA paralogs. Minor forms of miRNA/*s that are progressively away from predominant miRNA/* loci are not shown and can be referred to in Supplemental Tables 2 and 3. (c) In vitro DCL1 reconstitution assays with a synthetic TLBed pri-miRNA. Schematic illustration of the cleavage products by DCL1 is shown on right. Protein immunoprecipitation, cleavage assays and RNA processing were performed as described in Figure 2b. The positions of intact substrates, cleavage products, and RNA markers are shown. Red asterisks on transcripts indicate the 32P-labelling positions. Turquoise and purple arrows indicate abortive and productive processing sites, respectively.