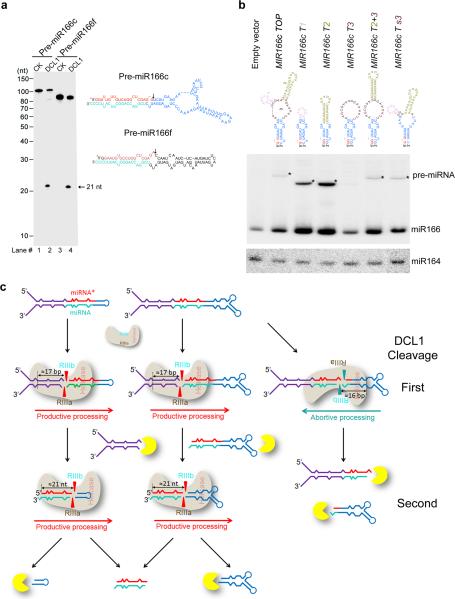
Figure 8.
Terminal loop affects steady-state abundance but not processing pattern of pre-miRNAs. (a) In vitro DCL1 reconstitution assays with the 5′ end labeled transcripts with pre-miR166c and pre-miR166f. Protein immunoprecipitation, cleavage assays, and processing of RNA products were performed as described in Figure 2b. The positions of intact substrates, cleavage products, and RNA markers are shown. Schematic illustration of the cleavage products by DCL1 is shown on right. Red asterisks on the transcripts indicate 32P-labelling positions. Black arrows show expected processing sites. (b) RNA blot analysis of pre-miR166 in the stable transformants expressing 35S-MIR166c mutants. RNA blot was probed using 32P-labelled oligo probes complementary to junction regions of miR166 and upper stem in pre-miR166c. Black triangles indicate pre-miRNAs. miR164 was probed as a loading control. (c) A model for bi-directional processing of pri-miRNA by DCL1. BTLs regulate miRNA biogenesis by triggering abortive processing of pri-miRNAs and destabilizing pre-miRNAs. Exo- or endo-ribonucleases are shown in yellow.