Figure 3. Generation and Validation of the Mtb Proteome Library.
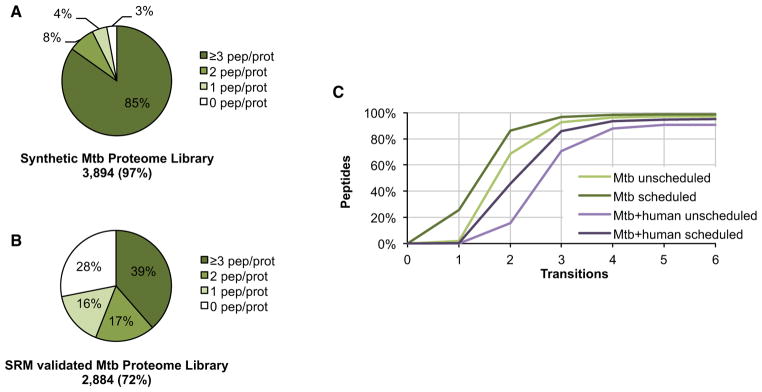
(A) Proteome coverage and distribution of the 15,679 synthetic peptides for which a fragment ion spectrum, and thus an SRM assay, could be obtained.
(B) Proteome coverage and distribution of the 7,094 peptides for which the synthetic SRM assay could be validated by SRM in a mixed unfractionated Mtb lysate of exponential and stationary phase cultures.
(C) Theoretical specificity of SRM assays determined by the SRMCollider algorithm is shown as a cumulative plot of the number of peptides which can be uniquely identified with a given number of transitions. Transitions were selected with decreasing intensity. Scheduled indicates that only background peptides with a retention time close to the target peptide are taken into consideration as interfering background.
See also Figures S1 and S2 and Table S1.
