Abstract
Periodontitis is an inflammatory disease caused by pathogenic microorganisms and characterized by the destruction of the periodontium. Obese individuals have an increased risk of periodontitis, and elevated circulating levels of adipokines, such as nicotinamide phosphoribosyltransferase (NAMPT), may be a pathomechanistic link between both diseases. The aim of this in vitro study was to examine the regulation of periodontal ligament (PDL) cells by NAMPT and its production under inflammatory and infectious conditions. NAMPT caused a significant upregulation of 9 genes and downregulation of 3 genes, as analyzed by microarray analysis. Eight of these genes could be confirmed by real-time PCR: NAMPT induced a significant upregulation of EGR1, MMP-1, SYT7, ITPKA, CCL2, NTM, IGF2BP3, and NRP1. NAMPT also increased significantly the MMP-1 and CCL2 protein synthesis. NAMPT was significantly induced by interleukin-1β and the periodontal microorganism P. gingivalis. NAMPT may contribute to periodontitis through upregulation of MMP-1 and CCL2 in PDL cells. Increased NAMPT levels, as found in obesity, may therefore represent a mechanism whereby obesity could confer an increased risk of periodontitis. Furthermore, microbial and inflammatory signals may enhance the NAMPT synthesis in PDL cells and thereby contribute to the increased gingival and serum levels of this adipokine, as found in periodontitis.
1. Introduction
Periodontitis is a chronic inflammatory disease, which is characterized by the destruction of the tooth-supporting tissues, such as the periodontal ligament (PDL), and caused by pathogenic microorganisms, such as Porphyromonas gingivalis, Tannerella forsythia, Treponema denticola, and Aggregatibacter actinomycetemcomitans. These periodontopathogens can trigger local production of proinflammatory mediators, such as interleukin-1β (IL-1β), cyclooxygenase 2 (COX2), as well as chemokine, cc motif, ligand 2 (CCL2), and matrix-degrading proteases, such as matrix metalloproteinase-1 (MMP-1), by infiltrating and resident cells of the periodontium. As a consequence of the exaggerated immunoinflammatory and proteolytic processes, the periodontal tissues are subjected to degradation and resorption, which can finally result in tooth loss [1, 2]. Data from the National Health and Nutrition Examination Survey III, which analyzed the health and nutritional status in the United States, demonstrate that approximately half of the US population aged ≥30 years is afflicted with periodontitis [3]. Periodontitis is also associated with systemic diseases and conditions, such as cardiovascular diseases, diabetes mellitus, and obesity, and has a negative influence on a wide range of physical, psychological, and social aspects of quality of life [4–6].
The exact mechanisms underlying the association between periodontitis and obesity are as yet unknown. However, it has been suggested that increased serum levels of adipokines derived from adipose tissue could make obese individuals more susceptible to periodontitis. The adipose tissue is not only an energy storage tissue but also acts as an endocrine organ, from which adipokines, such as nicotinamide phosphoribosyltransferase (NAMPT, also known as visfatin or pre-B-cell colony-enhancing factor 1), are secreted. Adipokines not only regulate insulin sensitivity and energy expenditure but also inflammatory and healing processes [7]. NAMPT is mainly produced by macrophages and adipocytes in the adipose tissue and induces production of inflammatory mediators and nuclear factor-kappaB (NFκB) activation [8]. High serum levels of NAMPT are found in obesity, metabolic syndrome, type 2 diabetes, atherosclerosis, and other diseases [9–11]. Therefore, increased serum levels of NAMPT could be at least one mechanism, whereby these diseases contribute to periodontitis. Recently, it has been demonstrated that NAMPT is also present in high levels in gingival crevicular fluid (GCF), gingival tissues, and serum from periodontally diseased patients, indicating that NAMPT might also be produced locally in the periodontium and play a role in the etiopathogenesis of periodontitis [12, 13]. However, whether NAMPT regulates the gene expression and protein synthesis of periodontal cells has yet to be elucidated. Furthermore, whether NAMPT is induced in these cells by periodontopathogens and IL-1β, which has been shown to be increased in GCF and gingival tissues at inflamed sites, is also largely unknown. This study was therefore established to examine the actions of NAMPT and its regulation by microbial and inflammatory signals in human PDL cells.
2. Materials and Methods
2.1. Culture and Treatment of Cells
PDL cells were harvested from periodontally healthy teeth that had to be extracted for orthodontic reasons. Written informed parental consent and approval of the Ethics Committee of the University of Bonn were obtained (#043/11). The cells were cultured in Dulbecco's minimal essential medium (DMEM, Invitrogen, Karlsruhe, Germany) supplemented with 10% fetal bovine serum (FBS, Invitrogen), 100 units penicillin, and 100 μg/mL streptomycin (Biochrom, Berlin, Germany) at 37°C in a humidified atmosphere of 5% CO2. PDL cells from passages 3 to 5 were seeded (50,000 cells/well) on cell culture plates and grown to 80% confluence. One day prior to the experiment, the FBS concentration was reduced to 1%. Medium was changed every other day.
In order to study the actions of NAMPT on PDL cells, various concentrations of NAMPT (30–300 ng/mL; Biomol, Hamburg, Germany) were added to cells. To mimic an inflammatory environment, cells were incubated with IL-1β (0.2–5 ng/mL; Calbiochem, San Diego, CA, USA), as done in our previous studies [14–16]. In order to simulate an infectious environment in vitro, cells were stimulated with the inactivated oral periodontopathogens Porphyromonas gingivalis ATCC 33277, Tannerella forsythia ATCC 43037, Treponema denticola ATCC 35405, and Aggregatibacter actinomycetemcomitans Y4 (optical density: 0.025, 0.05, and 0.1). Bacteria were suspended in PBS (OD660 nm = 1, equivalent to 1.2 × 109 bacterial cells/mL) and exposed two times to ultrasonication (160 W for 15 min) resulting in a complete killing. In the present study, cells were exposed to NAMPT, IL-1β, or periodontopathogens for up to 3 d. In order to unravel the intracellular mechanisms underlying the effects of NAMPT, PDL cells were preincubated with specific inhibitors against NFκB (pyrrolidine dithiocarbamate, PDTC; 10 μM; Calbiochem), JNK (SP600125; 10 μM; Calbiochem), p38 (SB203580; 10 μM; Calbiochem), MEK1/2 (U0126; 10 μM; Calbiochem), or PI3 K (50 μM; Calbiochem) signaling pathways 1 h before experiments.
2.2. Microarray Analysis
The regulatory effects of NAMPT on the gene expression of PDL cells were analyzed by a genomewide expression profiling using Illumina's HumanHT-12 v4 Expression BeadArrays (Illumina, San Diego, CA, USA). The RNA was reverse transcribed, amplified, and subsequently biotinylated using the Illumina TotalPrep-96 RNA Amplification Kit (Life Technologies, Carlsbad, CA, USA). The resulting cRNA was hybridized to the arrays according to the manufacturers manual using an automated liquid handling pipeline and scanned on an IScan System. The intensity signals were QC checked and exported using Illumina's GenomeStudio 2011 v1.1 software suite.
All array data underwent a rigorous quality control, before applying background correction (subtraction of an offset, which is estimated from the control probes) and a subsequent variance stabilizing transformation [17], followed by quantile normalization [18]. All steps were conducted using the lumi R-package [19]. After calculation of present/absent calls we only kept probes, which were present in each patient in each group. In order to consider additional confounding factors, which might potentially influence gene expression but are not directly captured by the available patient information, we applied the algorithm by Leek and Storey [20], which estimates a surrogate variable model. In addition, relative array quality weights were computed via the limma R-package [21] using the algorithm by Ritchie et al. [22]. Both, surrogate variables and array weights, can be considered when fitting a linear model to each gene via limma. In our case, the limma model contained two additive factors, namely, a donor and a group factor; that is, essentially we were fitting 2-way ANOVA model. After model fitting, differential gene expression for all clinically relevant differences can be extracted as contrasts from the model and corresponding P values can be computed (multiple testing correction) [23]. We refer the reader to the limma user guide for an excellent introduction [24].
2.3. Real-Time PCR
RNA was extracted using an RNA extraction kit (Qiagen, Hilden, Germany), and a total of 1 μg of RNA was reverse transcribed using iScriptTM Select cDNA Synthesis Kit (Bio-Rad Laboratories, Munich, Germany) at 42°C for 90 min followed by 85°C for 5 min. Expression of early growth response 1 (EGR1), MMP-1, synaptotagmin 7 (SYT7), inositol 1,4,5-trisphosphate 3-kinase a (ITPKA), CCL2, neurotrimin (NTM), insulin-like growth factor 2 mRNA-binding protein 3 (IGF2BP3), neuropilin 1 (NRP1), potassium channel tetramerization domain-containing protein 12 (KCTD12), heat-shock 27 kD protein 3 (HSPB3), transmembrane 4 L six family member 20 (TM4SF20), regulator of G protein signaling 4 (RGS4), NAMPT, COX2, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was detected by real-time PCR using the iCycler iQ detection system (Bio-Rad Laboratories), SYBR Green (Bio-Rad Laboratories), and specific primers (QuantiTect Primer Assay, Qiagen). One μL of cDNA was amplified as a template in a 25 μL reaction mixture containing 12.5 μL 2x QuantiFast SYBR Green PCR Master Mix (Qiagen), 2.5 μL of primers, and deionized water. The mixture was heated initially at 95°C for 5 min and then followed by 40 cycles with denaturation at 95°C for 10 s and combined annealing/extension at 60°C for 30 s. GAPDH was used as an endogenous control. The data were analyzed by the comparative threshold cycle (CT) method.
2.4. ELISA
The levels of NAMPT, MMP-1, and CCL2 in the supernatants of PDL cells were analysed by commercially available enzyme-linked immunoassay (ELISA) kits (RayBiotech, Norcross, GA, USA) according to the manufacturer's instructions. The absorbance was measured with a microplate reader (PowerWave x, BioTek Instruments, Winooski, VT, USA) at 450 nm. The data were normalized by the cell number, which was measured with an automatic cell counter (Moelab, Hilden, Germany).
2.5. Immunocytochemistry
PDL cells were cultured in the presence and absence of P. gingivalis ATCC 33277 or IL-1β on glass coverslips in 24-well plates. After 3 days, cells were fixed in 4% paraformaldehyde (Sigma-Aldrich, Munich, Germany) at pH 7.4 and room temperature (RT) for 15 min and then permeabilized in 0.1% Triton X-100 (Sigma-Aldrich) for 5 min. Non specific antigens were blocked by incubation with serum block (LSAB System; Santa Cruz Biotechnology, Santa Cruz, CA, USA) for 20 min. Subsequently, cells were incubated at RT for 90 min with rabbit polyclonal antibody to NAMPT (Santa Cruz Biotechnology). Afterwards cells were labeled with goat anti-rabbit IgG-HRP secondary antibody (Cell Signalling Technology, Danvers, MA, USA) for 45 min. For staining, cells were exposed to DAB chromogen (Dako, Hamburg, Germany) for 3 min at RT. After each incubation step, cells were washed twice with PBS (Sigma-Aldrich). Counterstaining was performed with Mayer's Haematoxylin (Merck Eurolab, Dietikon, Switzerland) for 2 min. Coverslips were mounted in DePex mounting medium (Serva Electrophoresis, Heidelberg, Germany). Standardized photomicrographs were taken using an Axioplan 2 imaging microscope (Carl Zeiss MicroImaging, Jena, Germany).
2.6. Immunofluorescence
PDL cells were fixed with 4% paraformaldehyde in PBS pH 7.4 for 10 min at RT, washed with PBS, and treated with 0.1% Triton X-100 for 5 min at RT. Then cells were washed again with PBS and blocked with a blocking buffer (nonfat dry milk; Bio-Rad Laboratories) for 1 h at RT. After washing, the cells were incubated with primary rabbit antibody NFκB p65 (1 : 400; Cell Signalling Technology) for 90 min and with secondary antibody CY3 (1 : 2,000; Abcam, Cambridge, MA, USA) for 45 min. Cells were observed under a 20x objective using an Axioplan 2 imaging microscope (Carl Zeiss MicroImaging). The images were captured with a PVCAM camera and the VisiView capturing software (Visitron Systems, Puchheim, Germany).
2.7. Statistical Analysis
The IBM SPSS Statistics 20 software was used for analysis. Mean values and standard errors of the mean (SEM) were calculated. All experiments were performed in triplicate and repeated at least twice. For statistical analysis, parametric (t test and ANOVA followed by the post-hoc Dunnett's test) and non parametric tests (Wilcoxon and Mann-Whitney U tests) were applied. Differences between groups were considered significant at P < 0.05. Microarray data were analyzed in the Bonn-Aachen International Center for IT, Algorithmic Bioinformatics, University of Bonn, Germany (see Section 2.2).
3. Results
3.1. Regulation of Gene Expression by NAMPT in PDL Cells
First, we sought to clarify whether NAMPT regulates the expression of genes in PDL cells by performing a microarray-based approach. As shown in Table 1, NAMPT caused a significant upregulation of 9 genes and a significant downregulation of 3 genes in PDL cells from 3 donors at 1 d. The NAMPT-upregulated genes were the following: EGR1, MMP-1, SYT7, ITPKA, CCL2, NTM, IGF2BP3, NRP1, and KCTD12. The 3 NAMPT-downregulated genes comprised HSPB3, TM4SF20, and RGS4 (Table 1). In PDL cells from 10 donors, the results from the microarray analysis were then validated by real-time PCR, which confirmed 8 out of the 12 NAMPT-regulated genes: NAMPT induced a significant upregulation of the mRNA expression for EGR1 (2.4-fold), MMP-1 (3.8-fold), SYT7 (4.6-fold), ITPKA (2.8-fold), CCL2 (2.3-fold), NTM (2.1-fold), IGF2BP3 (1.9-fold), and NRP1 (1.7-fold). KCTD12 was also upregulated by NAMPT, but the effect did not reach statistical significance (Table 2 and Figure 1(a)). The effects of NAMPT on the remaining genes could not be confirmed by PCR (data not shown).
Table 1.
Regulation of gene expression by NAMPT, as analyzed by microarray.
| Gene | LogFC | adj. P value |
|---|---|---|
| EGR1 | 2.184 | 0.008 |
| MMP-1 | 1.545 | 0.010 |
| SYT7 | 1.443 | 0.010 |
| ITPKA | 1.292 | 0.010 |
| CCL2 | 1.150 | 0.028 |
| NTM | 1.114 | 0.028 |
| IGF2BP3 | 1.040 | 0.020 |
| NRP1 | 1.024 | 0.010 |
| KCTD12 | 1.019 | 0.048 |
| HSPB3 | −1.035 | 0.030 |
| TM4SF20 | −1.164 | 0.028 |
| RGS4 | −1.397 | 0.025 |
Effect of NAMPT on gene expression of PDL cells from 3 donors at 1 d. Only genes with statistically significant (FDR < 5%) logFC ≥ 1 (upregulation by at least 2-fold) or logFC ≤ −1 (downregulation by at least 50%) were considered. Abbreviations: EGR1: early growth response 1; MMP-1: matrix metalloproteinase-1; SYT7: synaptotagmin 7; ITPKA: inositol 1,4,5-trisphosphate 3-kinase a; CCL2: chemokine, cc motif, ligand 2; NTM: neurotrimin; IGF2BP3: insulin-like growth factor 2 mRNA-binding protein 3; NRP1: neuropilin 1; KCTD12: potassium channel tetramerization domain-containing protein 12; HSPB3: heat-shock 27-kD protein 3; TM4SF20: transmembrane 4 L six family member 20; RGS4: regulator of G protein signaling 4.
Table 2.
Upregulation of gene expression by NAMPT, as analyzed by real-time PCR.
| Gene | Fold change (mean) |
SD | SEM | P value |
|---|---|---|---|---|
| EGR1 | 2.422 | 2.306 | 0.421 | <0.001 |
| MMP-1 | 3.781 | 3.116 | 0.569 | <0.001 |
| SYT7 | 4.575 | 7.947 | 1.451 | <0.001 |
| ITPKA | 2.764 | 3.182 | 0.581 | 0.001 |
| CCL2 | 2.285 | 1.346 | 0.246 | <0.001 |
| NTM | 2.124 | 2.460 | 0.449 | <0.001 |
| IGF2BP3 | 1.897 | 0.970 | 0.177 | <0.001 |
| NRP1 | 1.653 | 1.658 | 0.303 | <0.001 |
| KCTD12 | 1.713 | 0.958 | 0.175 | 0.095 |
Effect of NAMPT on gene expression of PDL cells from 10 donors at 1 d. Abbreviations: EGR1: early growth response 1; MMP-1: matrix metalloproteinase-1; SYT7: synaptotagmin 7; ITPKA: inositol 1,4,5-trisphosphate 3-kinase a; CCL2: chemokine, cc motif, ligand 2; NTM: neurotrimin; IGF2BP3: insulin-like growth factor 2 mRNA-binding protein 3; NRP1: neuropilin 1; KCTD12: potassium channel tetramerization domain-containing protein 12.
Figure 1.
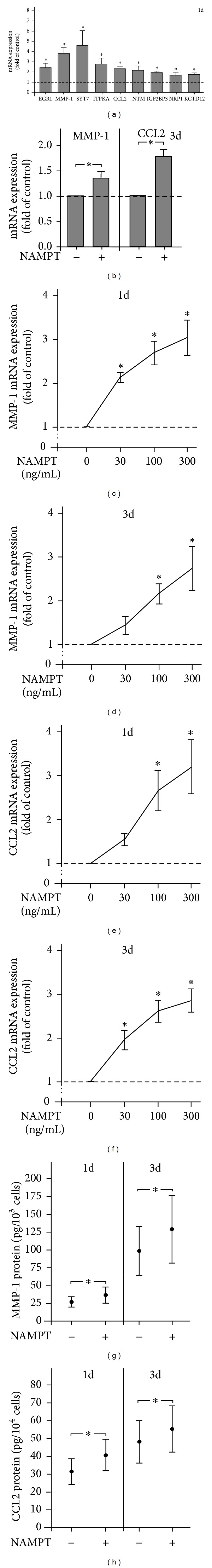
Stimulation of gene expression and protein synthesis by NAMPT. Upregulation of genes by NAMPT (100 ng/mL) in PDL cells from 10 donors at 1 d (a). Upregulation of MMP-1 and CCL2 expression by NAMPT (100 ng/mL) in PDL cells from 10 donors at 3 d (b). Stimulation of MMP-1 expression by various concentrations of NAMPT in PDL cells from 3 donors at 1 d (c) and 3 d (d). Stimulation of CCL2 expression by various concentrations of NAMPT in PDL cells from 3 donors at 1 d (e) and 3 d (f). Stimulation of MMP-1 protein synthesis by NAMPT (100 ng/mL) in PDL cells from 6 donors at 1 d and 3 d (g). Stimulation of CCL2 protein synthesis by NAMPT (100 ng/mL) in PDL cells from 6 donors at 1 d and 3 d (h). All experiments were performed in triplicate and repeated at least twice. Mean ± SEM; *significantly (P < 0.05) different from NAMPT-untreated cells (control).
3.2. Effect of NAMPT on MMP-1 and CCL2
Since MMP-1 and CCL2 are molecules known to be strongly associated with periodontitis, we further studied their regulation by NAMPT. As observed at 1 d, NAMPT increased significantly the MMP-1 (1.4-fold) and CCL2 (1.8-fold) expressions at 3 d (Figure 1(b)). Furthermore, the stimulatory effects of NAMPT on MMP-1 and CCL2 expressions were dose-dependent at both time points, as shown in Figures 1(c)–1(f). In addition, the NAMPT-induced upregulation of the MMP-1 and CCL2 expressions was parallelled by increased MMP-1 and CCL2 protein levels in the supernatants of NAMPT-stimulated cells, as compared to those of the control, at 1 d and 3 d (Figures 1(g) and 1(h)).
3.3. Exploitation of the JNK Pathway by NAMPT
Next, we studied how the effects of NAMPT on MMP-1 and CCL2 are mediated intracellularly. As evidenced by immunofluorescence microscopy, NAMPT stimulated the nuclear translocation of NFκB and caused a maximal NFκB accumulation within the nucleus at 60 min (Figure 2(a)). However, pre-incubation of cells with PDTC, a specific inhibitor of NFκB, had not effect on the NAMPT-stimulated upregulation of MMP-1 and CCL2 (data not shown). Moreover, the upregulation of both molecules by NAMPT was also not affected by inhibitors against the p38, MEK1/2 and PI3K signaling pathways. However, SP600125, an inhibitor of JNK signaling, blocked completely the stimulatory effect of NAMPT on the MMP-1 expression at 1 d (Figure 2(b)). In addition, SP600125 reduced significantly the NAMPT-stimulated upregulation of CCL2 by 69% at this time point (Figure 2(b)).
Figure 2.
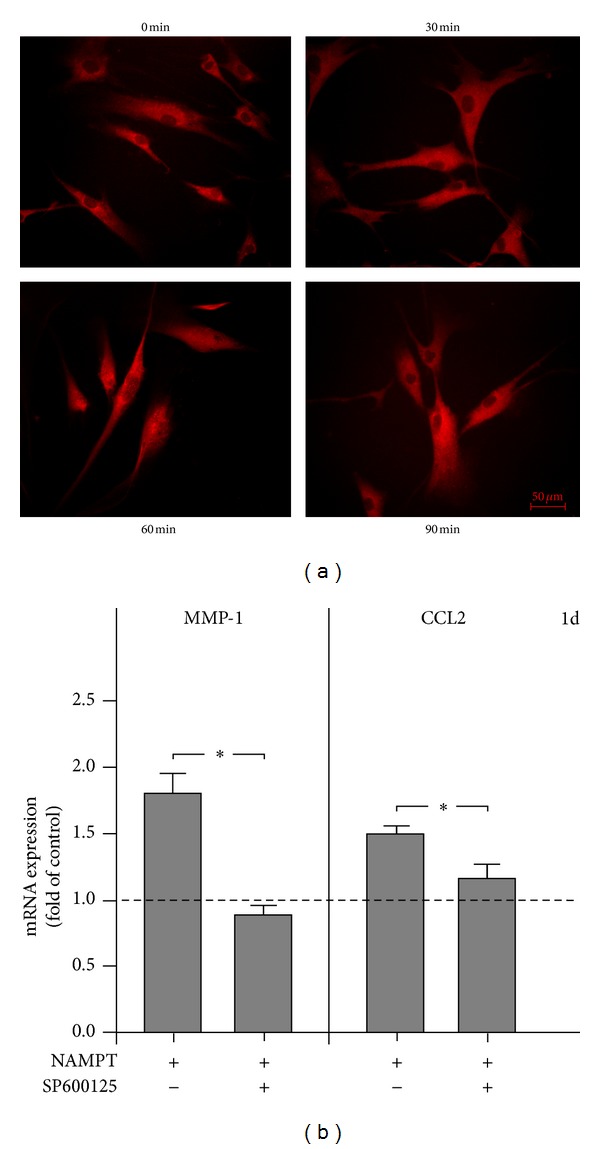
Intracellular pathways triggered by NAMPT. Stimulation of NFκB nuclear translocation by NAMPT at 60 min (a). Inhibition of the NAMPT-stimulated MMP-1 and CCL2 upregulation by preincubation of PDL cells with SP600125, a selective inhibitor of JNK, at 1 d (b). Images and data from one representative donor are shown. All experiments were performed in triplicate and repeated at least twice. Mean ± SEM; *significant (P < 0.05) difference between groups.
3.4. Stimulation of NAMPT Production by P. gingivalis
We then wondered whether pathogens that are strongly associated with periodontitis are capable of inducing the production of NAMPT in PDL cells. Whereas T. denticola, T. forsythia and A. actinomycetemcomitans had no significant effect, P. gingivalis increased significantly the NAMPT mRNA expression at 1 d and 3 d (Figures 3(a) and 3(b)). As shown in Figures 3(c) and 3(d), the actions of P. gingivalis on NAMPT mRNA expression were dose-dependent. Interestingly, pre-incubation with a specific inhibitor against NFκB signaling blocked completely the P. gingivalis-induced stimulation of NAMPT at 1 d. Furthermore, inhibitors against JNK and p38 signaling reduced the stimulatory effects of NAMPT by 87% and 37%, respectively, at this time point.
Figure 3.
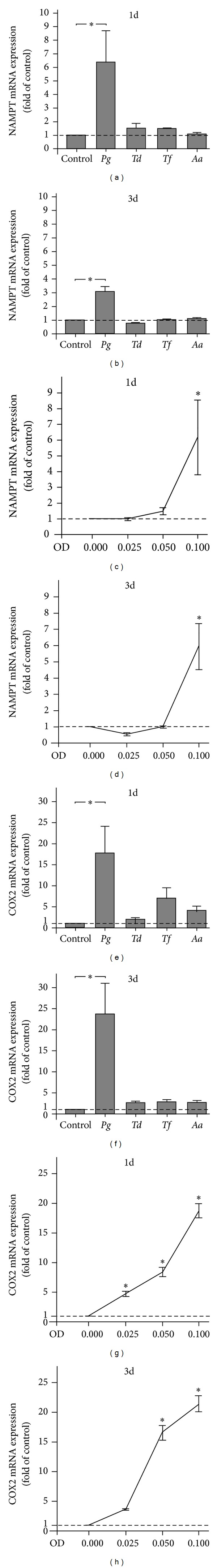
Regulation of NAMPT and COX2 expressions by periodontopathogens. NAMPT expression in PDL cells from 3 donors in response to various periodontopathogens (OD: 0.1; P. gingivalis, Pg; T. denticola, Td; T. forsythia, Tf; A. actinomycetemcomitans, Aa) at 1 d (a) and 3 d (b). Stimulation of NAMPT expression in PDL cells from 3 donors by various concentrations of P. gingivalis (OD: 0.025, 0.050, and 0.100) at 1 d (c) and 3 d (d). COX2 expression in PDL cells from 3 donors in response to various periodontopathogens (OD: 0.1; P. gingivalis, Pg; T. denticola, Td; T. forsythia, Tf; A. actinomycetemcomitans, Aa) at 1 d (e) and 3 d (f). Stimulation of COX2 expression in PDL cells from 3 donors by various concentrations of P. gingivalis (OD: 0.025, 0.050, and 0.100) at 1 d (g) and 3 d (h). All experiments were performed in triplicate and repeated at least twice. Mean ± SEM; *significantly (P < 0.05) different from P. gingivalis-untreated cells (control).
The stimulation of NAMPT expression by P. gingivalis was also observed at protein level, as analyzed by ELISA: at 1 d, PDL cells in the presence and absence of P. gingivalis produced 28.83 ± 0.29 ng NAMPT protein/104 cells and 17.91 ± 0.58 ng NAMPT protein/104 cells, respectively. At 3 d, P. gingivalis-stimulated cells and control cells synthesized 30.97 ± 3.09 ng NAMPT protein/104 cells and 21.65 ± 2.94 ng NAMPT protein/104 cells, respectively. The differences between both groups at each time point were significant. The stimulation of NAMPT protein production by P. gingivalis was also evidenced by immunocytochemistry (Figure 4(c)).
Figure 4.
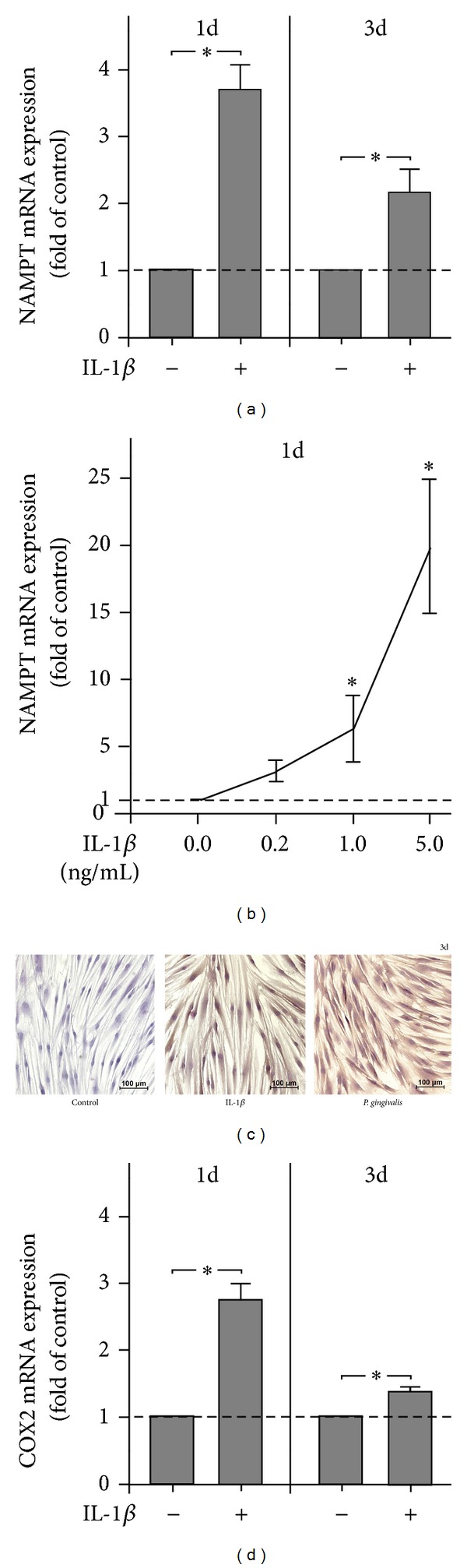
Regulation of NAMPT and COX2 expressions by IL-1β. NAMPT expression in PDL cells from 6 donors in response to IL-1β (1 ng/mL) at 1 d and 3 d (a). Stimulation of NAMPT expression in PDL cells from 3 donors by various concentrations of IL-1β (0.2, 1.0, and 5.0 ng/mL) at 1 d (b). NAMPT protein synthesis in PDL cells in the presence and absence of IL-1β and P. gingivalis at 3 d. Images from one representative donor are shown (c). COX2 expression in PDL cells from 6 donors in response to IL-1β (1 ng/mL) at 1 d and 3 d (d). All experiments were performed in triplicate and repeated at least twice. Mean ± SEM; *significantly (P < 0.05) different from IL-1β-untreated cells (control).
Like NAMPT, COX2 was significantly upregulated by P. gingivalis at 1 d and 3 d (Figures 3(e) and 3(f)). By contrast, all other periodontopathogens were not capable of stimulating the COX2 expression at both time points (Figures 3(e) and 3(f)). The P. gingivalis-induced stimulation of COX2 expression was dose-dependent (Figures 3(g) and 3(h)).
3.5. Stimulation of NAMPT Production by IL-1β
Finally, we examined whether NAMPT is also induced by IL-1β in PDL cells. Interestingly, IL-1β upregulated significantly the NAMPT expression at 1 d and 3 d (Figure 4(a)). Moreover, the actions of IL-1β on NAMPT expression were dose-dependent at 1 d (Figure 1(b)) but not at 3 d (data not shown). When cells were preincubated with an inhibitor against the MEK1/2 signaling pathway, the IL-1β-induced stimulation of NAMPT expression was completely suppressed. Moreover, inhibitors against JNK, NFκB, and p38 signaling reduced the stimulatory effect of IL-1β on NAMPT by 80%, 78%, and 46%, respectively.
The upregulation of NAMPT at transcriptional level was paralleled by increased NAMPT protein levels in the supernatants of IL-1β-stimulated cells: at 1 d, PDL cells in the presence and absence of IL-1β synthesized 32.68 ± 3.62 ng NAMPT protein/104 cells and 20.28 ± 4.36 ng NAMPT protein/104 cells, respectively. At 3 d, IL-1β-treated cells and control cells produced 30.97 ± 3.82 ng NAMPT protein/104 cells and 20.43 ± 0.76 ng NAMPT protein/104 cells, respectively. The differences between groups were significant at both time points. The stimulation of NAMPT protein synthesis by IL-1β was also observed by immunocytochemistry (Figure 4(c)).
IL-1β also induced a significant upregulation of COX2 expression at 1 d and 3 d (Figure 4(d)).
4. Discussion
Our study shows for this first time that NAMPT stimulates the production of CCL2 and MMP-1 in human PDL cells, suggesting that NAMPT may contribute to periodontal inflammation and matrix destruction through the production of these molecules. Furthermore, NAMPT is induced by the periodontopathogen P. gingivalis and the proinflammatory cytokine IL-1β in PDL cells, which shows that microbial and inflammatory signals may use this adipokine for their detrimental effects on the periodontium.
MMP-1 degrades specifically type I collagen and, additionally, types II, III, V, IX, and X collagen, thereby playing a critical role for modeling and remodeling of the extracellular matrix [25]. Several studies have revealed that MMP-1 levels are increased in GCF and human gingiva from periodontitis patients [26, 27]. Furthermore, MMP-1 levels could be significantly reduced by periodontal treatment [27]. CCL2 is synthesized by a variety of cell types and regulates the migration and infiltration of monocytes, memory T lymphocytes, and natural killer cells. In addition, CCL2 seem to influence T-cell immunity [28]. Higher levels of CCL2 in GCF and gingiva from inflamed sites have been reported [20–31]. Our experiments provide original evidence that NAMPT induces increased expression and synthesis of MMP-1 and CCL2 in human PDL cells, as evidenced by several assays and in a high number of donors. These findings are in accordance with previous observations in nonperiodontal cells and underline the proinflammatory and catabolic role of NAMPT in the pathophysiology of periodontitis [32–34]. Since increased NAMPT levels are found in obesity, our data suggest at least one mechanism whereby obesity could confer an increased risk of periodontitis in obese individuals [5, 9].
Surprisingly, NAMPT also upregulated a number of genes, most of which have mainly been reported in oncology and neurosciences. The transcription factor EGR1 is a regulator of multiple tumor suppressors but, paradoxically, can also promote tumor progression [35, 36]. EGR1 is altered by hypoxia [37], stimulates angiogenesis [35], and regulates the immune system [38]. Synaptotagmins are transmembrane proteins with two Ca2+-binding C(2) domains in their cytosolic region. Syt7 regulates the exocytosis of lysosomes and thereby plays a role in bone resorption [39], cell migration [40], CTL responses [41], and neurodegeneration [42]. ITPKA phosphorylates inositol 1,4,5-trisphosphate, thereby modulating the calcium (Ca2+) level within the cell and the levels of a large number of inositol polyphosphates [43]. ITPKA promotes cell motility and the metastatic potential of tumor cells [43, 44]. Furthermore, ITPKA is critical for the spatial and temporal regulation of spine actin remodeling, synaptic plasticity, and learning and memory [45]. Little is known about neurotrimin, a neural cell adhesion protein. It has been shown to be an estrogen-regulated determinant of peripheral sympathetic innervation [46] and to play a role in axonal fasciculation of specific cerebellar systems and the formation of excitatory synapses and their stabilization [47]. The IGF2BPs bind to the 5′-untranslated region of IGF-2 mRNA and regulate a number of important aspects of cell function, such as cell polarization, morphology, migration, proliferation, differentiation, and invasion [48, 49]. NRP1 plays a critical part in neuronal development, in angiogenesis and tumor invasion [50, 51]. NRP1 is a coreceptor for members of the vascular endothelial growth factor family and the class 3 semaphorins, which are polypeptides with roles in axonal guidance [52]. Furthermore, NRP1 mediates interaction of regulatory T cells with dendritic cells and modulates the immune response [53]. KCTDs act as auxiliary subunits of GABA(B) receptors and generate desensitizing receptor responses [54]. Further studies should clarify the physiological and pathological role of these genes in the periodontium.
We also sought to examine the intracellular mechanism whereby the stimulatory effects of NAMPT on MMP-1 and CCL2 are accomplished. Although NAMPT stimulated the NFκB nuclear translocation in PDL cells, which concurs with observations in other cells [32, 55, 56], pre-incubation of cells with a specific NFκB inhibitor did not abrogate the NAMPT actions on MMP-1 and CCL2 in our cells. However, the NFκB signaling pathway could be involved in the NAMPT-induced upregulation of some of the other genes which were identified by microarray and PCR analyses. Interestingly, our experiments revealed that the NAMPT-induced upregulation of MMP-1 and CCL2 was JNK dependent, which is supported by findings from others [32].
Recently, increased NAMPT levels have been found in GCF, gingival tissues, and serum from periodontally diseased patients, as compared to periodontally healthy individuals. This suggests that NAMPT might also be produced locally in the periodontium and regulated by periodontopathogens and/or inflammatory mediators [12, 13]. Interestingly, we found that P. gingivalis, a key pathogen associated with periodontitis, and the proinflammatory cytokine IL-1β, which is increased at inflamed periodontal sites, can induce NAMPT in PDL cells, thereby supporting the assumption that NAMPT is locally produced in the presence of an infectious and/or inflammatory environment. By upregulation of MMP-1 and CCL2, NAMPT may mediate and amplify proinflammatory and proteolytic actions of P. gingivalis and IL-1β on the PDL. The finding that NAMPT is induced by P. gingivalis is in line with our previous experiments, which have demonstrated that NAMPT is upregulated by F. nucleatum, a gram-negative, anaerobic microorganism, which acts as a bridge bacterium between early and late colonizers during plaque development [57]. Interestingly, T. forsythia, T. denticola, and A. actinomycetemcomitans did not regulate the NAMPT expression, at least at the concentration tested, in the present study. Our observation that NAMPT is increased by IL-1β in PDL cells concurs with findings in other cells, which have also been shown to produce increased NAMPT levels in response to IL-1β [58–60]. Our data provide evidence that PDL cells produce increased levels of NAMPT under infectious and inflammatory conditions, which suggests that local production of NAMPT in the inflamed periodontium could contribute to the increased gingival and serum levels of NAMPT, as observed in patients with periodontitis. NAMPT could therefore also represent a pathomechanistic link, how periodontitis affects systemic diseases, such as diabetes mellitus and cardiovascular diseases.
Interestingly, the upregulation of NAMPT by P. gingivalis could be completely blocked by an inhibitor against NFκB and reduced by inhibitors against the JNK and p38 signaling pathways. Recently, other investigators have also shown that P. gingivalis induces NFκB activation and p38 signaling for its actions [61, 62]. An inhibitor against MEK1/2 signaling pathway suppressed completely the IL-1β-induced stimulation of NAMPT expression. JNK, NFκB, and p38 signaling were also found to be involved in the stimulatory effects of IL-1β on NAMPT. That IL-1β exploits the NFκB and mitogen-activated protein kinase pathways for its actions is well known and supported by the present findings [63, 64].
In order to simulate an infectious environment, PDL cells were incubated with a suspension of various periodontopathogens. Since the suspensions were exposed to intensive ultrasonication, the suspensions contained disrupted cell wall particles with a high amount of lipopolysaccharide. However, other bacterial components may also have been present in the suspension. The concentrations used in this study were determined by dose-response experiments. However, a significant upregulation of NAMPT was only observed for P. gingivalis. P. gingivalis is a gram-negative bacterium and strongly associated with periodontitis. For its detrimental effects, P. gingivalis possesses a number of virulence factors, such as gingipains and fimbriae. P. gingivalis can invade host cells and also evade the host defense system [65–67]. As it has been shown in several cells that COX2, an enzyme which is responsible for the formation of prostanoids, is upregulated by P. gingivalis and IL-1β, we also analyzed the COX2 expression as a positive control in our study [61, 62, 68, 69]. As expected, P. gingivalis and IL-1β increased the COX2 expression in a dose- and time-dependent manner in PDL cells.
In summary, the present study demonstrates for the first time that NAMPT stimulates the production of MMP-1 and CCL2 in human PDL cells, which suggests that NAMPT may contribute to periodontal inflammation and matrix destruction through the production of these molecules. Therefore, increased NAMPT levels, as found in obesity, may represent at least one mechanism, whereby obesity could confer an increased risk of periodontitis in obese individuals. In addition, our study provides original evidence that NAMPT is induced by the periodontopathogen P. gingivalis and the proinflammatory cytokine IL-1β in PDL cells, which shows that microbial and inflammatory signals may use this adipokine for their detrimental effects on the periodontium. These findings suggest that local production of NAMPT in the inflamed PDL could contribute to the increased gingival and serum levels of NAMPT, as observed in patients with periodontitis and therefore represent a pathomechanistic link whereby periodontitis affects systemic diseases, such as diabetes mellitus and cardiovascular diseases.
Conflict of Interests
The authors declare that they have no conflict of interests.
Acknowledgments
The authors would like to thank Ms. Ramona Hömig, Professor Werner Götz, and Professor Stephan Baader for their support. This study was funded by the German Research Foundation (KFO208/TP4) and the University of Bonn. Professor Holger Fröhlich is a member of the Exzellenzcluster ImmunoSensation.
References
- 1.Pihlstrom BL, Michalowicz BS, Johnson NW. Periodontal diseases. The Lancet. 2005;366(9499):1809–1820. doi: 10.1016/S0140-6736(05)67728-8. [DOI] [PubMed] [Google Scholar]
- 2.Sbordone L, Bortolaia C. Oral microbial biofilms and plaque-related diseases: microbial communities and their role in the shift from oral health to disease. Clinical Oral Investigations. 2003;7(4):181–188. doi: 10.1007/s00784-003-0236-1. [DOI] [PubMed] [Google Scholar]
- 3.Albandar JM. Commentary: underestimation of periodontitis in nhanes surveys. Journal of Periodontology. 2011;82(3):337–341. doi: 10.1902/jop.2011.100638. [DOI] [PubMed] [Google Scholar]
- 4.Beikler T, Flemmig TF. Oral biofilm-associated diseases: trends and implications for quality of life, systemic health and expenditures. Periodontology 2000. 2011;55(1):87–103. doi: 10.1111/j.1600-0757.2010.00360.x. [DOI] [PubMed] [Google Scholar]
- 5.Suvan J, D’Aiuto F, Moles DR, Petrie A, Donos N. Association between overweight/obesity and periodontitis in adults. A systematic review. Obesity Reviews. 2011;12(501):e381–e404. doi: 10.1111/j.1467-789X.2010.00808.x. [DOI] [PubMed] [Google Scholar]
- 6.Seymour GJ, Ford PJ, Cullinan MP, Leishman S, Yamazaki K. Relationship between periodontal infections and systemic disease. Clinical Microbiology and Infection. 2007;13(4):3–10. doi: 10.1111/j.1469-0691.2007.01798.x. [DOI] [PubMed] [Google Scholar]
- 7.Conde J, Scotece M, Gómez R, et al. Adipokines: biofactors from white adipose tissue. A complex hub among inflammation, metabolism, and immunity. BioFactors. 2011;37(6):413–420. doi: 10.1002/biof.185. [DOI] [PubMed] [Google Scholar]
- 8.Moschen AR, Gerner RR, Tilg H. Pre-b cell colony enhancing factor/nampt/visfatin in inflammation and obesityrelated disorders. Current Pharmaceutical Design. 2010;16(17):1913–1920. doi: 10.2174/138161210791208947. [DOI] [PubMed] [Google Scholar]
- 9.Chang Y-H, Chang D-M, Lin K-C, Shin S-J, Lee Y-J. Visfatin in overweight/obesity, type 2 diabetes mellitus, insulin resistance, metabolic syndrome and cardiovascular diseases: a meta-analysis and systemic review. Diabetes/Metabolism Research and Reviews. 2011;27(6):515–527. doi: 10.1002/dmrr.1201. [DOI] [PubMed] [Google Scholar]
- 10.Taskesen D, Kirel B, Us T. Serum visfatin levels, adiposity and glucose metabolism in obese adolescents. Journal of Clinical Research in Pediatric Endocrinology. 2012;4:76–81. doi: 10.4274/Jcrpe.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang LQ, Heruth DP, Ye SQ. Nicotinamide phosphoribosyltransferase in human diseases. Journal of Bioanalysis and Biomedicine. 2011;3(1):13–25. doi: 10.4172/1948-593X.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pradeep AR, Raghavendra N.M. RNM, Prasad MVR, Kathariya R, Patel SP, Sharma A. Gingival crevicular fluid and serum visfatin concentration: their relationship in periodontal health and disease. Journal of Periodontology. 2011;82(9):1314–1319. doi: 10.1902/jop.2011.100690. [DOI] [PubMed] [Google Scholar]
- 13.Pradeep AR, Raghavendra NM, Sharma A, et al. Association of serum and crevicular visfatin levels in periodontal health and disease with type 2 diabetes mellitus. Journal of Periodontology. 2012;83(5):629–634. doi: 10.1902/jop.2011.110272. [DOI] [PubMed] [Google Scholar]
- 14.Nokhbehsaim M, Deschner B, Winter J, et al. Interactions of regenerative, inflammatory and biomechanical signals on bone morphogenetic protein-2 in periodontal ligament cells. Journal of Periodontal Research. 2011;46(3):374–381. doi: 10.1111/j.1600-0765.2011.01357.x. [DOI] [PubMed] [Google Scholar]
- 15.Nokhbehsaim M, Winter J, Rath B, Jäger A, Jepsen S, Deschner J. Effects of enamel matrix derivative on periodontal wound healing in an inflammatory environment in vitro. Journal of Clinical Periodontology. 2011;38(5):479–490. doi: 10.1111/j.1600-051X.2010.01696.x. [DOI] [PubMed] [Google Scholar]
- 16.Nokhbehsaim M, Deschner B, Winter J, et al. Anti-inflammatory effects of EMD in the presence of biomechanical loading and interleukin-1β in vitro. Clinical Oral Investigations. 2012;16(1):275–283. doi: 10.1007/s00784-010-0505-8. [DOI] [PubMed] [Google Scholar]
- 17.Lin SM, Du P, Huber W, Kibbe WA. Model-based variance-stabilizing transformation for Illumina microarray data. Nucleic Acids Research. 2008;36(2, article e11) doi: 10.1093/nar/gkm1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bolstad BM, Irizarry RA, Åstrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 19.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24(13):1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 20.Leek JT, Storey JD. A general framework for multiple testing dependence. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(48):18718–18723. doi: 10.1073/pnas.0808709105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statistical Applications in Genetics and Molecular Biology. 2004;3(1, article 3) doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 22.Ritchie ME, Diyagama D, Neilson J, et al. Empirical array quality weights in the analysis of microarray data. BMC Bioinformatics. 2006;7, article 261 doi: 10.1186/1471-2105-7-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society B. 1995;57:89–300. [Google Scholar]
- 24.Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey V, Huber W, Irizarry R, Dudoit S, editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. New York, NY, USA: Springer; 2005. pp. 397–420. [Google Scholar]
- 25.Arakaki PA, Marques MR, Santos MCLG. MMP-1 polymorphism and its relationship to pathological processes. Journal of Biosciences. 2009;34(2):313–320. doi: 10.1007/s12038-009-0035-1. [DOI] [PubMed] [Google Scholar]
- 26.Ejeil A-L, Igondjo-Tchen S, Ghomrasseni S, Pellat B, Godeau G, Gogly B. Expression of matrix metalloproteinases (MMPs) and tissue inhibitors of metalloproteinases (TIMPs) in healthy and diseased human gingiva. Journal of Periodontology. 2003;74(2):188–195. doi: 10.1902/jop.2003.74.2.188. [DOI] [PubMed] [Google Scholar]
- 27.Tüter G, Kurtiş B, Serdar M. Effects of phase I periodontal treatment on gingival crevicular fluid levels of matrix metalloproteinase-1 and tissue inhibitor of metalloproteinase-1. Journal of Periodontology. 2002;73(5):487–493. doi: 10.1902/jop.2002.73.5.487. [DOI] [PubMed] [Google Scholar]
- 28.Deshmane SL, Kremlev S, Amini S, Sawaya BE. Monocyte chemoattractant protein-1 (MCP-1): an overview. Journal of Interferon and Cytokine Research. 2009;29(6):313–326. doi: 10.1089/jir.2008.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garlet GP, Martins W, Jr., Ferreira BR, Milanezi CM, Silva JS. Patterns of chemokines and chemokine receptors expression in different forms of human periodontal disease. Journal of Periodontal Research. 2003;38(2):210–217. doi: 10.1034/j.1600-0765.2003.02012.x. [DOI] [PubMed] [Google Scholar]
- 30.Gupta M, Chaturvedi R, Jain A. Role of monocyte chemoattractant protein-1 (MCP-1) as an immune-diagnostic biomarker in the pathogenesis of chronic periodontal disease. Cytokine. 2013;61:892–897. doi: 10.1016/j.cyto.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 31.Kurtiş B, Tüter G, Serdar M, et al. Gingival crevicular fluid levels of monocyte chemoattractant protein-1 and tumor necrosis factor-alpha in patients with chronic and aggressive periodontitis. Journal of Periodontology. 2005;76(11):1849–1855. doi: 10.1902/jop.2005.76.11.1849. [DOI] [PubMed] [Google Scholar]
- 32.Brentano F, Schorr O, Ospelt C, et al. Pre-B cell colony-enhancing factor/visfatin, a new marker of inflammation in rheumatoid arthritis with proinflammatory and matrix-degrading activities. Arthritis and Rheumatism. 2007;56(9):2829–2839. doi: 10.1002/art.22833. [DOI] [PubMed] [Google Scholar]
- 33.Liu SW, Qiao SB, Yuan JS, Liu DQ. Visfatin stimulates production of monocyte chemotactic protein-1 and interleukin-6 in human vein umbilical endothelial cells. Hormone and Metabolic Research. 2009;41(4):281–286. doi: 10.1055/s-0028-1102914. [DOI] [PubMed] [Google Scholar]
- 34.Sommer G, Kralisch S, Kloting N, et al. Visfatin is a positive regulator of MCP-1 in human adipocytes in vitro and in mice in vivo. Obesity. 2010;18(8):1486–1492. doi: 10.1038/oby.2009.462. [DOI] [PubMed] [Google Scholar]
- 35.Adamson ED, Mercola D. Egr1 transcription factor: multiple roles in prostate tumor cell growth and survival. Tumor Biology. 2002;23(2):93–102. doi: 10.1159/000059711. [DOI] [PubMed] [Google Scholar]
- 36.Baron V, Adamson ED, Calogero A, Ragona G, Mercola D. The transcription factor Egr1 is a direct regulator of multiple tumor suppressors including TGFβ1, PTEN, p53, and fibronectin. Cancer Gene Therapy. 2006;13(2):115–124. doi: 10.1038/sj.cgt.7700896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ten VS, Pinsky DJ. Endothelial response to hypoxia: physiologic adaptation and pathologic dysfunction. Current Opinion in Critical Care. 2002;8(3):242–250. doi: 10.1097/00075198-200206000-00008. [DOI] [PubMed] [Google Scholar]
- 38.McMahon SB, Monroe JG. The role of early growth response gene 1 (egr-1) in regulation of the immune response. Journal of Leukocyte Biology. 1996;60(2):159–166. doi: 10.1002/jlb.60.2.159. [DOI] [PubMed] [Google Scholar]
- 39.Teitelbaum SL. The osteoclast and its unique cytoskeleton. Annals of the New York Academy of Sciences. 2011;1240(1):14–17. doi: 10.1111/j.1749-6632.2011.06283.x. [DOI] [PubMed] [Google Scholar]
- 40.Colvin RA, Means TK, Diefenbach TJ, et al. Synaptotagmin-mediated vesicle fusion regulates cell migration. Nature Immunology. 2010;11(6):495–502. doi: 10.1038/ni.1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fowler KT, Andrews NW, Huleatt JW. Expression and function of synaptotagmin VII in CTLs. Journal of Immunology. 2007;178(3):1498–1504. doi: 10.4049/jimmunol.178.3.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Glavan G, Schliebs R, Živin M. Synaptotagmins in neurodegeneration. Anatomical Record. 2009;292(12):1849–1862. doi: 10.1002/ar.21026. [DOI] [PubMed] [Google Scholar]
- 43.Kato H, Uzawa K, Onda T, et al. Down-regulation of 1D-myo-inositol 1,4,5-trisphosphate 3-kinase A protein expression in oral squamous cell carcinoma. International Journal of Oncology. 2006;28(4):873–881. [PubMed] [Google Scholar]
- 44.Windhorst S, Fliegert R, Blechner C, et al. Inositol 1,4,5-trisphosphate 3-kinase-A is a new cell motility-promoting protein that increases the metastatic potential of tumor cells by two functional activities. Journal of Biological Chemistry. 2010;285(8):5541–5554. doi: 10.1074/jbc.M109.047050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Il HK, Soon KP, Soon TH, et al. Inositol 1,4,5-trisphosphate 3-kinase A functions as a scaffold for synaptic rac signaling. Journal of Neuroscience. 2009;29(44):14039–14049. doi: 10.1523/JNEUROSCI.2483-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krizsan-Agbas D, Pedchenko T, Smith PG. Neurotrimin is an estrogen-regulated determinant of peripheral sympathetic innervation. Journal of Neuroscience Research. 2008;86(14):3086–3095. doi: 10.1002/jnr.21768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen S, Gil O, Ren YQ, Zanazzi G, Salzer JL, Hillman DE. Neurotrimin expression during cerebellar development suggests roles in axon fasciculation and synaptogenesis. Journal of Neurocytology. 2002;30(11):927–937. doi: 10.1023/a:1020673318536. [DOI] [PubMed] [Google Scholar]
- 48.Bell JL, Wächter K, Mühleck B, et al. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): post-transcriptional drivers of cancer progression? Cellular and Molecular Life Sciences. 2013;70(15):2657–2675. doi: 10.1007/s00018-012-1186-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suvasini R, Shruti B, Thota B, et al. Insulin growth factor-2 binding protein 3 (IGF2BP3) is a glioblastoma-specific marker that activates phosphatidylinositol 3-kinase/mitogen-activated protein kinase (PI3K/MAPK) pathways by modulating IGF-2. Journal of Biological Chemistry. 2011;286(29):25882–25890. doi: 10.1074/jbc.M110.178012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Staton CA, Kumar I, Reed MWR, Brown NJ. Neuropilins in physiological and pathological angiogenesis. Journal of Pathology. 2007;212(3):237–248. doi: 10.1002/path.2182. [DOI] [PubMed] [Google Scholar]
- 51.Wild JRL, Staton CA, Chapple K, Corfe BM. Neuropilins: expression and roles in the epithelium. International Journal of Experimental Pathology. 2012;93(2):81–103. doi: 10.1111/j.1365-2613.2012.00810.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zachary IC. How neuropilin-1 regulates receptor tyrosine kinase signalling: the knowns and known unknowns. Biochemical Society Transactions. 2011;39(6):1583–1591. doi: 10.1042/BST20110697. [DOI] [PubMed] [Google Scholar]
- 53.Mizui M, Kikutani H. Neuropilin-1: the glue between regulatory T cells and dendritic cells? Immunity. 2008;28(3):302–303. doi: 10.1016/j.immuni.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 54.Schwenk J, Metz M, Zolles G, et al. Native GABAB receptors are heteromultimers with a family of auxiliary subunits. Nature. 2010;465(7295):231–235. doi: 10.1038/nature08964. [DOI] [PubMed] [Google Scholar]
- 55.Adya R, Tan BK, Chen J, Randeva HS. Nuclear factor-κb induction by visfatin in human vascular endothelial cells. Diabetes Care. 2008;31(4):758–760. doi: 10.2337/dc07-1544. [DOI] [PubMed] [Google Scholar]
- 56.Fan Y, Meng S, Wang Y, Cao J, Wang C. Visfatin/PBEF/Nampt induces EMMPRIN and MMP-9 production in macrophages via the NAMPT-MAPK (p38, ERK1/2)-NF-κB signaling pathway. International Journal of Molecular Medicine. 2011;27(4):607–615. doi: 10.3892/ijmm.2011.621. [DOI] [PubMed] [Google Scholar]
- 57.Nogueira AV, Nokhbehsaim M, Eick S, et al. Regulation of visfatin by microbial and biomechanical signals in PDL cells. doi: 10.1007/s00784-013-0935-1. Clinical Oral Investigations. In press. [DOI] [PubMed] [Google Scholar]
- 58.Gosset M, Berenbaum F, Salvat C, et al. Crucial role of visfatin/pre-B cell colony-enhancing factor in matrix degradation and prostaglandin E2 synthesis in chondrocytes: possible influence on osteoarthritis. Arthritis and Rheumatism. 2008;58(5):1399–1409. doi: 10.1002/art.23431. [DOI] [PubMed] [Google Scholar]
- 59.Kendal CE, Bryant-Greenwood GD. Pre-B-cell colony-enhancing factor (PBEF/Visfatin) gene expression is modulated by NF-κB and AP-1 in human amniotic epithelial cells. Placenta. 2007;28(4):305–314. doi: 10.1016/j.placenta.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 60.Williams MR, Kataoka N, Sakurai Y, Powers CM, Eskin SG, McIntire LV. Gene expression of endothelial cells due to interleukin-1 beta stimulation and neutrophil transmigration. Endothelium. 2008;15(1-2):73–84. doi: 10.1080/10623320802092443. [DOI] [PubMed] [Google Scholar]
- 61.Murakami Y, Kawata A, Seki Y, et al. Comparative inhibitory effects of magnolol, honokiol, eugenol and bis-eugenol on cyclooxygenase-2 expression and nuclear factor-kappa B activation in RAW264. 7 macrophage-like cells stimulated with fimbriae of Porphyromonas gingivalis. In Vivo. 2012;26:941–950. [PubMed] [Google Scholar]
- 62.Reddi D, Brown SJ, Belibasakis GN. Porphyromonas gingivalis induces RANKL in bone marrow stromal cells: involvement of the p38 MAPK. Microbial Pathogenesis. 2011;51(6):415–420. doi: 10.1016/j.micpath.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 63.Dunne A, O’Neill LAJ. The interleukin-1 receptor/Toll-like receptor superfamily: signal transduction during inflammation and host defense. Science"s STKE. 2003;2003(171, article re3) doi: 10.1126/stke.2003.171.re3. [DOI] [PubMed] [Google Scholar]
- 64.Lee J-K, Kim S-H, Lewis EC, Azam T, Reznikov LL, Dinarello CA. Differences in signaling pathways by IL-1β and IL-18. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(23):8815–8820. doi: 10.1073/pnas.0402800101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Holt SC, Ebersole JL. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the “red complex”, a prototype polybacterial pathogenic consortium in periodontitis. Periodontology 2000. 2005;38:72–122. doi: 10.1111/j.1600-0757.2005.00113.x. [DOI] [PubMed] [Google Scholar]
- 66.Nakayama K. Molecular genetics of Porphyromonas gingivalis: gingipains and other virulence factors. Current Protein and Peptide Science. 2003;4(6):389–395. doi: 10.2174/1389203033486983. [DOI] [PubMed] [Google Scholar]
- 67.Potempa J, Sroka A, Imamura T, Travis J. Gingipains, the major cysteine proteinases and virulence factors of Porphyromonas gingivalis: structure, function and assembly of multidomain protein complexes. Current Protein and Peptide Science. 2003;4(6):397–407. doi: 10.2174/1389203033487036. [DOI] [PubMed] [Google Scholar]
- 68.Chang Y-C, Huang F-M, Yang S-F, et al. Induction of cyclooxygenase-2 mRNA and protein expression in human pulp cells stimulated with black-pigmented bacteroides. Journal of Endodontics. 2003;29(4):240–243. doi: 10.1097/00004770-200304000-00002. [DOI] [PubMed] [Google Scholar]
- 69.Hayashi Y, Kobayashi M, Kuwata H, et al. Interferon-γ and interleukin 4 inhibit interleukin 1β-induced delayed prostagilandin E2 generation through suppression of cyclooxugenase-2 expression in human fibroblasts. Cytokine. 2000;12(6):603–612. doi: 10.1006/cyto.1999.0622. [DOI] [PubMed] [Google Scholar]


