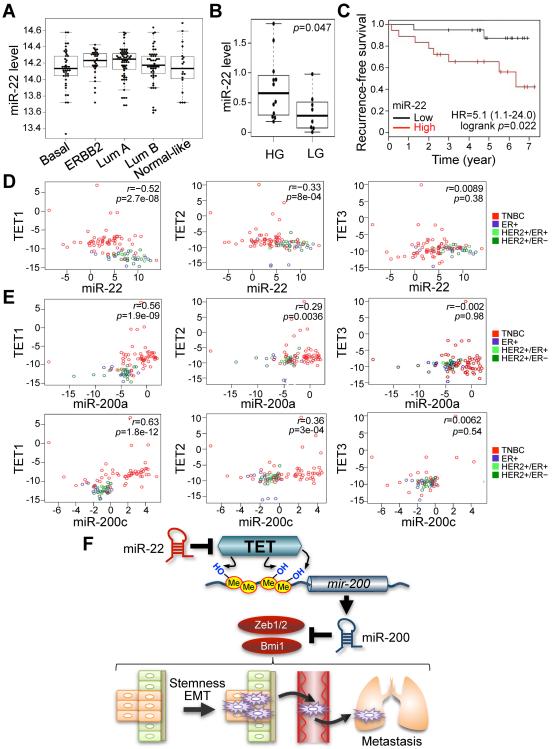
Figure 7. miR-22 overexpression correlates with poor clinical outcomes and silencing of the TET-miR-200 axis in patients.
(A) miR-22 expression profiling was analyzed from a previously published Illumina Human RefSeq-8 and miRNAv1 array dataset (superSeries GSE22220). Breast tumors were classified by molecular subtypes, estrogen receptor (ER), progesterone receptor (PR), ERBB2 (HER-2) and epithelial clusters.
(B) miR-22 expression was analyzed by using a real-time qPCR with RNAs from human breast cancer patient samples. Tumor grade was determined by the parameters, ER, PR, ERBB2 (HER-2) and epithelial clusters. HG, ER-positive high-grade tumors; LG, ER-positive low-grade tumors.
(C) Non triple-negative breast cancer (TNBC) patient samples were sub-divided into two groups according to low and high expression of miR-22 with median split of all samples. A Kaplan-Meier plot representing the disease-free survival of patients was stratified.
(D) Anti-correlation between miR-22 and TET family expressions was analyzed using a real-time qPCR with RNAs from breast cancer patient samples.
(E) Co-expression analysis of TET family and miR-200a/miR-200c was analyzed by using a real-time qPCR with RNAs from breast cancer patient samples.
(F) Proposed model of the role of miR-22 for EMT, stemness and metastasis through epigenetic inactivation of miR-200 by directly targeting the TET family. miR-22 decreases the level of 5hmC by negatively regulating TET family, followed by epigenetic inactivation of miR-200 due to the reduced 5hmC levels. Ultimately, dysfunction of miR-200 triggers EMT and stemness, which in turn increases mammary tumorigenesis and metastasis.