Figure 10.
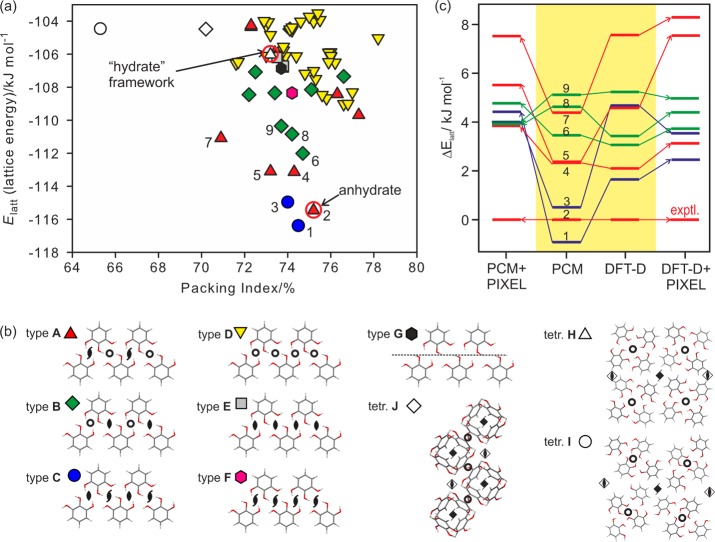
(a) Lattice energy landscape for pyrogallol anhydrate (Elatt = Uinter + ΔEintra, PCM). Each symbol denotes a crystal structure, which is a lattice energy minimum classified by the most extensive common-packing motif based on the hydrogen bonding shown in (b). (c) Relative lattice energies of the most stable computationally generated pyrogallol structures calculated using different methods: PCM, isolated molecule relaxed structures with average polarization from the PCM model as in (a), DFT-D, periodic density functional theory relaxations with dispersion correction, and PIXEL calculations using either the PCM or DFT-D optimized structure. Tie lines have been added to show the changes in relative ordering. The numbers labeling the symbols in (a) identify the crystal structures by stability order using the PCM model (Table S9a of the Supporting Information). Only selected symmetry operations are drawn in (b).