Fig. 3.
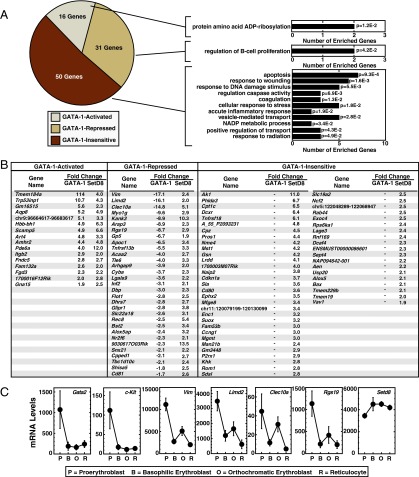
SetD8 as a context-dependent GATA-1 corepressor. The set of genes up-regulated by SetD8 knockdown was subjected to sorting into one of three categories based on their response to GATA-1 activation: GATA-1–activated, GATA-1–repressed, or GATA-1–insensitive. GATA-1–sensitivity values are based on microarray analysis comparing cells receiving control siRNA, and either no treatment or β-estradiol (n = 3 individual experiments). (A) Pie chart displays the percentage of SetD8 regulated genes that fall into each of the three categories. GO analysis was conducted with all gene categories (David Bioinformatics, sorted by P value). (B) Genes falling into each category are listed. (C) mRNA levels of GATA-1–repressed genes mined from the murine ErythronDB database (www.cbil.upenn.edu/ErythronDB/). The direct target genes Gata2 and c-Kit are shown as controls, and Vim, Clec10a, and Rgs19 represent SetD8/GATA-1–corepressed genes.