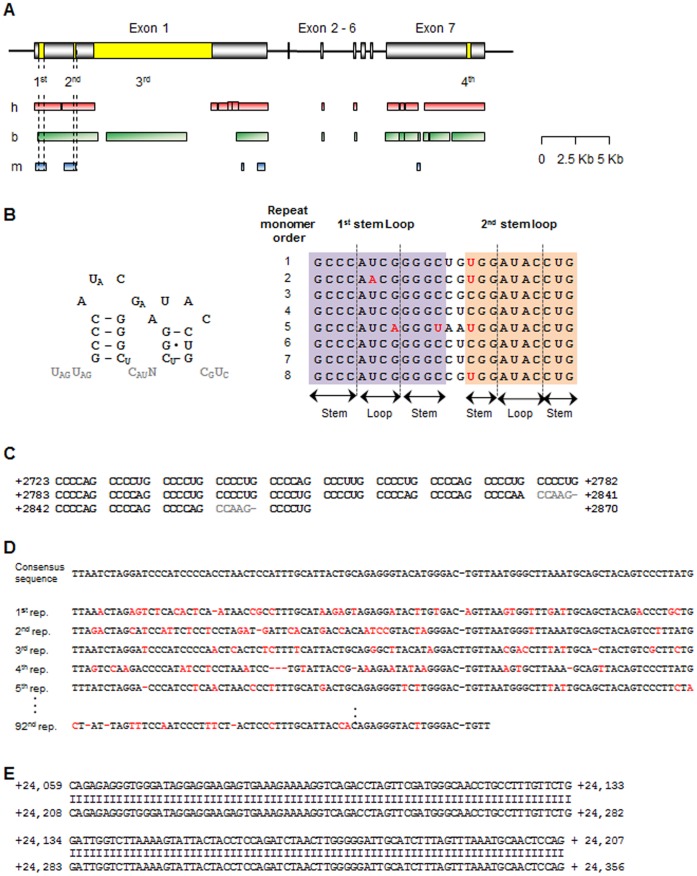
Figure 2. Analysis of repeat sequences in porcine XIST RNA.
(A) Distribution of repeated sequences in the porcine XIST gene model. Gray rectangles indicate exons, and the yellow boxes in the exons indicate repeat regions. The regions having homology with human, bovine, and mouse XIST/Xist are represented to red (h), green (b), and blue (m) boxes, respectively. The first and second repeat regions showed similarity with all accessed XIST/Xist sequence from other species (dashed lines). The diagram is scaled. (B) The first repeat region in porcine XIST. A monomer of the first repeat region in pigs (left panel) share a consensus sequence and a predicted secondary structure of the mouse [28]. Gray text indicates non-conserved sequences. Eight repeated monomers identified in the porcine XIST first repeat region (+327 to +695) were aligned with a consensus sequence (Right panel). Red character indicates different sequence compared to the consensus sequence in conserved stem-loop region. (C) The second repeat region (+2,723 to +2,870) is shown, with non-repeat sequences in gray. (D) The third repeat region (+4,302 to +13,102) fraction. The first five repeats and last repeat were aligned with the consensus sequence. Red characters mean the sequences which were not same to consensus sequence. (E) Alignment of the fourth repeat region (+24,059 to +24,356). Two copies of the monomer were perfectly matched.