Abstract
The citrus mealybug, Planococcus citri, is an important plant pest with a very broad plant host range. P. citri is a phloem feeder and loss of plant vigor and stunting are characteristic symptoms induced on a range of host plants, but P. citri also reduces fruit quality and causes fruit drop leading to significant yield reductions. Better strategies for managing this pest are greatly needed. RNA interference (RNAi) is an emerging tool for functional genomics studies and is being investigated as a practical tool for highly targeted insect control. Here we investigated whether RNAi effects can be induced in P. citri and whether candidate mRNAs could be identified as possible targets for RNAi-based P. citri control. RNAi effects were induced in P. citri, as demonstrated by specific target reductions of P. citri actin, chitin synthase 1 and V-ATPase mRNAs after injection of the corresponding specific double-stranded RNA inducers. We also used recombinant Tobacco mosaic virus (TMV) to express these RNAi effectors in Nicotiana benthamiana plants. We found that P. citri showed lower fecundity and pronounced death of crawlers after feeding on recombinant TMV-infected plants. Taken together, our data show that actin, chitin synthase 1 and V-ATPase mRNAs are potential targets for RNAi against P. citri, and that recombinant TMV is an effective tool for evaluating candidate RNAi effectors in plants.
Introduction
The citrus mealybug, Planococcus citri (Hemiptera: Pseudococcidae), is a serious pest of citrus, nursery plants, grapes and is occasionally considered a serious pest of greenhouse crops [1]. On citrus, P. citri damages plants by feeding on roots, bark, foliage and fruit [2]. During feeding P. citri injects toxic saliva into plants and extracts phloem sap, and induces symptoms including defoliation, fruit discoloration and fruit drop [2]. In addition, P. citri secretes copious amounts of honeydew leading to the development of sooty mold which also leads to reduced fruit quality and lowered plant vigor through the loss of photosynthetic capacity.
RNA interference (RNAi) is a natural gene regulation and anti-viral defense found in eukaryotes. RNAi has not only been used to study gene function [3]–[5], but it has shown as a potential strategy for controlling insect pests [6]–[9]. Double-stranded (ds) and short-interfering RNAs (siRNAs) both can induce RNAi effects in insects [10], resulting in degradation of the target mRNA. If the target mRNA is critical for insect development, severe phenotypic effects or even death may result [10], [11]. RNAi effects have been achieved for insects by introducing specific ds- and siRNAs into the insects body by microinjection [12], [13], by feeding [14]–[20] or even by exogenous applications [21]. For plant-feeding insects, using transgenic plants engineered to express dsRNAs corresponding to insect target mRNAs has also shown potential [7], [17]. Thus, for practical RNAi application it is critical to identify targets which are vital for the insect and which could successfully be suppressed through RNAi.
RNAi effects have been demonstrated in many insects including fruit flies (Drosophila melanogaster), flour beetles (T. castaneum), pea aphids (Acyrthosiphon pisum), and tobacco hornworms (Manduca sexta) that were all selectively killed when fed species-specific dsRNAs targeting V-ATPase mRNAs [10], [18]. Moreover, other studies have shown that using a transient plant-mediated RNAi delivery system for pea aphid (A. pisum) against the rak1 gene that is predominantly expressed in the gut, results in great reduction of the rak1 mRNA levels. This approach was further extended to the C002 gene that is expressed in the salivary glands and has also shown reduction in the mRNA levels of C002 [16]. In another study on potato/tomato psyllids (Bactericerca cockerelli) gut-specific mRNAs were successfully targeted through microinjection and oral acquisition of interfering RNAs [10]. Recently, RNAi effects were induced in the Glassy-winged sharpshooter (Homalodisca vitripennis) by microinjection of interfering RNAs corresponding to actin mRNAs, thus reducing the mRNA levels and causing mortality in the injected H. vitripennis [11]. Furthermore, RNAi has been used successfully in transgenic corn plants against Diabrotica virgifera virgifera [7]. These transgenic plants showed a reduction in damage caused by D. virgifera virgifera suggesting that the RNAi pathway can be exploited to control insect pests via in planta expression of interfering RNAs. Earlier studies also showed reductions of CYP6AE14 transcripts in the D. virgifera virgifera midgut after feeding the larvae on plant material expressing dsRNAs specific to CYP6AE14, and this also resulted in a severe retardation of larval growth [17].
By contrast RNAi effects were not successfully achieved for several other insects. In Bombyx mori, injection of dsRNA targeting the ecdysone receptor mRNA or other targets in the epidermis did not result in detectable RNAi effects [22]. Similarly, a series of unsuccessful attempts through feeding dsRNAs were reported for Helicoverpa armigera and Spodoptera frugiperda [22], while two other independent studies showed efficient targeting of mRNAs expressed in the S. frugiperda midgut and brain after feeding dsRNAs [23], [24]. Interestingly, while feeding long dsRNAs to H. armigera larvae was not very successful [22], good RNAi effects were achieved using custom-designed siRNAs [25], suggesting different efficiencies for short siRNAs vs. long dsRNAs, or at least their specific sequences, in these species. In general, there seems to be a positive correlation between amount of dsRNA and the degree of RNAi effects [22]. Interestingly, for Bicyclus anynana, Chrysodeixis includes and Spodoptera littoralis [22], [26], [27] even higher amounts of dsRNA (>1 mg per mg of tissue) did not result in detectable RNAi effects. This variation in responses to RNAi by different species may be due to variation in delivery methods, dose of effector RNAs introduced, RNAi targets or by differences in species with respect to their susceptibility to RNAi [22].
The above studies suggest that RNAi approaches could potentially be used as a tailor-made insecticide for at least some insects, but the correct target, effector RNA, and means of effector delivery must be identified in order to achieve desired RNAi effects. It is also likely that in insects, a continuous supply of effector RNAs may be required. Most insects appear to lack RdRp (RNA dependent RNA Polymerase) [6], [28]–[30] which in plants serves to amplify the RNAi effects thereby producing a continuous supply of effector RNAi molecules [31]. Thus, transgenic plants engineered to express a continuous supply of RNAi effectors are desirable. However, all mRNAs are not equal targets, and assessing potential effector RNAs via transgenic plants is a tedious, time consuming and expensive effort [32]. Recombinant plant virus vectors engineered to express the desired insect effector RNAs in plants provide a potential alternative approach that circumvents many of the above problems seen for transgenic plants [32]. One such virus is Tobacco mosaic virus (TMV) [33]. TMV has a wide plant host range, it is easily engineered, mechanically transmissible from plant to plant, and plants respond quickly to virus infection by producing TMV-specific siRNAs. Then if TMV can be engineered to contain sequences of the desired mRNA target, siRNAs towards this target will be generated. Furthermore, TMV replicates through dsRNA intermediates, and thus plants infected with recombinant TMVs produce at least two forms of RNAi effectors, specific siRNAs, and replicative form dsRNAs.
There is no information available on RNAi activity in P. citri. In this study we first determined if RNAi effects could be induced in P. citri and then we attempted to identify potential mRNA targets that might offer opportunities for P. citri control using RNAi based approaches. We used in vitro synthesized dsRNAs and recombinant-TMV infected plants and demonstrated the effectiveness of P. citri RNAi targets via both of these assays.
Materials and Methods
Insect rearing
Insects were collected from naturally infested Vitis vinifera plants on the U. C. Davis campus with the permission of greenhouse manager Mr. Steven Silva. P. citri were reared on Nicotiana tabacum and Solanum lycopersicum plants under greenhouse conditions at 24–28°C. Second instar P. citri nymphs were used in feeding and 3rd instars for injection bioassay studies to evaluate the RNAi effects.
RNA extractions
RNA was extracted from whole insects or insect body parts using Trizol reagent following the manufacturer's protocol (Ambion, Cat. No. 15596018). RNAs were precipitated using two volumes of ethanol and 1/10 volume of 3 M sodium acetate. RNA pellets were washed with 70% ethanol, and re-suspended in DEPC-treated diH2O.
PCR amplification and analysis of P. citri sequences
Published chitin synthase 1 (CHS1) sequences of Aedes aegypti (XM_001662150.1), Anopheles gambiae (XM_321951.2), Ostrinia furnacalis (EU258740.1), Culex quinquefasciatus (XM_001864559.1), Plutella xylostella (AB271784.1), Nasonia vitripennis (XM_001602131.2) and Tribolium castaneum (NM_001039403.1) were aligned using ClastalW. Universal forward and reverse primers (Table 1) were designed from conserved regions to amplify a 507bp fragment of CHS1 from P. citri. V-ATPase primers were designed in the same fashion from the conserved region of the gene by aligning published V-ATPase sequences of B. mori (NM_001098359.1), M. sexta (X64233.1), Apis mellifera (XM_623492.3), T. castaneum (XM_971095.2), A. aegypti (AF008922.1), Aedes albopictus (AY864912.1) and N. vitripennis (XM_003426538.1). Actin gene sequence was isolated using primers designed from P. solenopsis actin gene sequence (unpublished data) (Table 1).
Table 1. Primers used for RT-PCR and cDNA synthesis.
| SequenceName | Primer Name | Primer sequence 5′ to 3′ | Product Size (bp) | RT-PCR Conditions |
| Chitin synthase | CSFCSR | GGCTCTGGTCCTATGGTGTGGTATCA aGCCACRAAAGCACCCACCAACATAAGGAA | 507 | 94°C for 3min, 35cycles of 94°C for 30Sec, 48°C for 45sec, 72°C for 1min and finally 72°C for 10 min |
| V-ATPase | VATPase F1VATPase R1 | aGGCYACYATHCAGGTATAaTCVARMCCCCAGAACAC | 1122 | 94°C for 3min, 35cycles of 94°C for 30Sec, 50°C for 45sec, 72°C for 1min and finally 72°C for 10 min |
| Actin | MbActinSalFMbActinSmaR | bGTCGACTCCGGTGATGGTGTATCTCAcCCCGGGATCACGGACGATTTCTCGTT | 171 | 94°C for 3min, 35cycles of 94°C for 30Sec, 50°C for 45sec, 72°C for 1min and finally 72°C for 10 min |
| PJL36 | PJL36 leftPJL36 Right | AGATCTTACAGTATCACTACTCC GTACGCACCACGTGTGATTACGG | 100 | 94°C for 3min, 35cycles of 94°C for 30Sec, 55°C for 45sec, 72°C for 1min and finally 72°C for 10 min |
The following degeneracy codes are used in the primer sequences:
R = G/A, Y = C/T, H = A/C/T, V = A/C/G, M = A/C.
Underlined nucleotide sequence is the Sal 1 restriction enzyme recognition sequence, added to facilitate cloning.
Underlined sequence is the Sma 1 restriction enzyme recognition sequence, added to facilitate cloning.
First strand cDNA was synthesized using the superscript cDNA synthesis kit and with an oligo (dT)-primer using 500 ng RNA and following manufacturer's instruction (Invitrogen, Cat. No. 11904–018). Briefly RNA was incubated with oligo (dT) primer at 65°C for 5 min. A 10 µl reaction mixture (10X RT buffer 2 µl, 25 mM MgCl2 4 µl, 0.1 M DTT 2 µl, RNAse out (40 U/µl) 1 µl and SuperScript II 1 µl) was added and samples were incubated at 42°C for 50 min followed by 10 min incubation at 70°C. The cDNA (200 ng/reaction) was used as template for amplification through PCR using GoTaq Flexi DNA polymerase (Promega Corp., Madison, WI, USA Cat. No. M8291) and universal primers at the PCR conditions given (Table 1).
PCR products were analyzed by agarose gel electrophoresis, purified using the ZymoClean PCR cleaning kit (Zymo Corp., Irvine, CA, USA Cat. No. D4006), and subsequently cloned into pGEM-T Easy (Promega Corp., Madison, WI, USA Cat. No. A1360). Sequencing was done by the DNA sequencing facility at UC Davis using M13 forward and reverse primers. The resulting sequences were confirmed by protein translations and BLAST searches (http://www.ncbi.nlm.nih.gov) and gene-specific primers were designed. A second round of PCRs was performed to amplify the cDNAs to be used to generate dsRNAs (Table 2). The PCR products were sequenced and sequences were submitted to GenBank (NCBI) with accession numbers JX443530 and JX443529 of CHS1 and V-ATPase gene fragments, respectively. The actin sequence was submitted to European Nucleotide Archive (EMBL) with accession number HF952554.
Table 2. Primers used for RT-PCR of RNAi targets.
| SequenceName | Primer Name | Primer sequence 5′ to 3′ | Product Size (bp) | RT-PCR Conditions |
| Chitin synthase | PcCHSSalFPcCHSSmaR | aCGTCGAC CGTTGGCTGTGCACATTACT bCCCGGG AAGCACCCCACCAACATAAG | 290 | 94°C for 3min, 35cycles of 94°C for 30Sec, 55°C for 45sec, 72°C for 1min and finally 72°C for 10 min |
| V-ATPase | PCVATPSalFPCVATPSmaR | aCGTCGACAAGGTTGGCAGCCACATAACbCCCGGGCGAAAGCTCCTGGTATAGCG | 332 | 94°C for 3min, 35cycles of 94°C for 30Sec, 57°C for 45sec, 72°C for 1min and finally 72°C for 10 min |
| Actin | PCActinSalFPCActinSmaR | aCGTCGACTCCGGTGATGGTGTATCTCAbCCCGGGATTTCTCGTTCGGCAGTTGT | 168 | 94°C for 3min, 35cycles of 94°C for 30Sec, 55°C for 45sec, 72°C for 1min and finally 72°C for 10 min |
| GFP | PacI-GFP leftJAL34 | cCGGTTAATTAAATGGCTAGCAAAGGAGAAGAACdTTTGCGGCCGCTTATTTGTAGAGCTCATCCATG | 742 | 94°C for 3min, 35cycles of 94°C for 30Sec, 55°C for 45sec, 72°C for 1min and finally 72°C for 10 min |
Underlined nucleotide sequence is the Sal 1 restriction enzyme recognition sequence, added to facilitate cloning.
Underlined sequence is the Sma 1 restriction enzyme recognition sequence, added to facilitate cloning.
Underlined nucleotide sequence is the Pac 1 restriction enzyme recognition sequence, added to facilitate cloning.
Underlined nucleotide sequence is the Not 1 restriction enzyme recognition sequence, added to facilitate cloning.
DsRNA synthesis and microinjection of P. citri
The RNAi target regions of CHS1 and V-ATPase mRNAs used here were selected using the SnapDragon dsRNA design software (http://www.flyrnai.org/cgi-bin/RNAi_find_primers.pl). Regions with sufficient nucleotide divergence from those of other insect species were selected to avoid potential off-target effects. The T7 promoter sequence was added to the 5′ ends of both forward and reverse primers (Table 3), PCR products were amplified from cDNA and purified using the ZymoClean PCR cleaning kit (Zymo Corp., Irvine, CA, USA Cat. No: D4006). Purified PCR products were used as templates to synthesize dsRNA using T7 RNA polymerase and the MEGAscript RNAi Kit as per manufacturer's instructions (Ambion, Austin, TX, USA Cat. No. AM1626). The dsRNAs were treated with DNAse and RNAse at 37°C for 30 min following the manufacturer's protocol, and electrophoresed on an agarose gel to assess dsRNA quality.
Table 3. Primers used for in vitro dsRNA synthesis.
| Sequence Name | Primer Name | aPrimer sequence 5′ to 3′ | Product Size (bp) |
| Chitin Synthase | PCCHST7FPCCHST7R | GGATCCTAATACGACTCACTATAGGG CGTTGGCTGTGCACATTACT GGATCCTAATACGACTCACTATAGGG AAGCACCCCACCAACATAAG | 342 |
| V-ATPase | PCVATPT7FPCVATPT7R | GGATCCTAATACGACTCACTATAGGG AAGGTTGGCAGCCACATAAC GGATCCTAATACGACTCACTATAGGG CGAAAGCTCCTGGTATAGCG | 384 |
| Actin | PCActinT7FPCActinT7R | GGATCCTAATACGACTCACTATAGGG TCCGGTGATGGTGTATCTCA GGATCCTAATACGACTCACTATAGGG ATTTCTCGTTCGGCAGTTGT | 220 |
| GFP | GFPFGFPR | bCTAATACGACTCACTATAGG GCGGCCGCACGCGTGCTGAAGTCAAGTTbCTAATACGACTCACTATAGG GCGGCCGCCTTTTCGTTGGGATCTTTCG | 321 |
Underlined nucleotide sequence is the T7 RNA polymerase promoter sequence. A pair of gene specific primers with the T7 promoter sequence at the 5′ ends of both primers was used to amplify the desired templates. Amplified PCR products were used as templates for in vitro dsRNA synthesis.
Italic nucleotide sequence is the Not 1 restriction enzyme recognition sequence.
To assess if RNAi effects could be induced in P. citri, individual mealybug third instar nymphs were injected with 200 nl of dsRNA by using a micromanipulator and microinjector (http://www.tritechresearch.com/MINJ-PD.html). For microinjection, insects were placed individually on a 2% agarose gel to immobilize them, and were injected with 200 nl (500 ng/µL) of dsRNA using fine glass needles and the micromanipulator/miroinjector. Injected insects were moved back to fresh tobacco leaves at 24–26oC for post-injection observations and procedures. RNA was extracted from injected P. citri at 48 h, 72 h, 96 h and 150 h post-injection and qRT-PCR was performed to assess target mRNA amounts.
Cloning in TMV
Cloned sequences for the mRNA targets of interest (actin, CHS1 and V-ATPase) were excised from pGEM-T Easy by digesting with NotI, and ligated into the NotI-digested TMV-based vector, pJL36 [33]. Positive clones were screened first by restriction digestion and then by nucleotide sequence analysis to confirm the insert sequence orientation. Confirmed clones were transformed into Agrobacterium tumefaciens (GV3101 strain) for inoculation to N. benthamiana plants. Plants were inoculated by agro infiltration with the recombinant TMV-based vector following the protocol as described previously [34]. RT-PCR was performed using insect gene specific primers to check the presence of insect target mRNA in the inoculated plants. The pJL24 TMV-based vector has been engineered to express green fluorescent protein (GFP) was used as a control. Reverse transcriptase polymerase chain reaction (RT-PCR) was used to verify retention of the inserted sequence by the recombinant TMVs in infected plants. RNA was extracted from upper symptomatic leaves of inoculated plants, and primers from the TMV vector sequence flanking the insert were used for RT-PCR (Table 1).
SiRNA detection
Seven days post inoculation total RNAs were extracted from plant leaves using Trizol reagent following the manufacturer's instructions. The small RNA fraction was isolated from total RNA using 50% PEG 8000 precipitation. Small RNAs were electrophoresed on a 15% 8 M urea polyacrylamide gel with microRNA markers (New England BioLabs, Inc, Ipswich, MA, USA Cat. No. N2102S). RNAs were transferred to Hybond-NX Amersham membrane (GE Healthcare, Piscata way, NJ Cat. No. RPN203T) using a semi dry blotter as described [35]. A 32P-UTP labeled probe was prepared by in vitro transcription using the Maxiscript T7 kit (Ambion, Austin, TX, USA Cat. No. AM1312) as described [11] and the resulting probe was fragmented by adding carbonation buffer (20 mM Na2CO3 and 80mM NaHCO3). Hybridization was done overnight in Ultrahyb-oligo hybridization buffer (Ambion, Austin, TX, USA Cat. No. AM8669) at 42°C followed by two washes in Northern Max Low Stringency Wash Solution (Ambion, Austin, TX, USA Cat. No. AM8673) for 15 minutes at 42°C. The blot was drained and then exposed to X-Ray film (F-BX810, Phenix) at −80°C for 72 hrs.
Insect bioassays and Quantitative Real-time PCR (qRT-PCR) analyses
At seven days post inoculation (dpi), ten 2nd instar mealybugs were placed on inoculated and upper leaves of N. benthamiana plants. Data were recorded on mortality, insect growth, ovisac production, crawler emergence and their survival. RNA was extracted from individual mealybugs after feeding for 12 days on the respective TMV-infected plants, and four days post dsRNA-injection. cDNA was synthesized using 500 ng total RNA and oligo(dT) primer. Quantitative real time polymerase chain reaction (qRT-PCR) was performed using gene specific primers with the 18S rRNA as the internal control (Table 4) using Fast SYBR Green Master Mix (Applied Biosystem Cat. No. 4385610) and comparative mRNA amounts were analyzed by 2–ΔΔCT method [10] using ABI 7500 system software v2.0.1. The reaction mixture contained 8.5 µl SYBR Green Master Mix, 0.5 µl 10 µM forward primer, 0.5 µl 10 µM reverse primer and 1 µl cDNA template in a final reaction volume of 20 µl. Forty cycles of PCR were performed for 3 sec at 95oC and 30 sec at 60oC. The ΔCt value for individual samples were obtained by subtracting average Ct (cycle threshold) value for 18S rRNA from average Ct value for the target RNA. One of the samples in control groups (elution buffer (EB) injected in dsRNA microinjection experiments and GFP dsRNA or siRNA-fed mealybugs in feeding experiments) was selected as a reference sample. The ΔΔCt value for individual samples was obtained by subtracting ΔCt value for the reference sample from the ΔCt value of the test sample. The relative expression of target RNA in comparison to 18S RNA level in each sample is represented by the 2–ΔΔCt value. The mean 2–ΔΔCt value of all samples was divided with the mean 2–ΔΔCt value of the control sample to make easy comparison between the groups. The average value and standard deviation value (SD value) of the 2–ΔΔCt values in treatment and control groups were calculated separately, and the student's t-test was performed between treated and control groups to see the difference between the groups.
Table 4. Primers used for qRT-PCR.
| Sequence Name | Primer Name | Primer sequence 5′ to 3′ |
| Chitin synthase | PCCHSRTFPCCHSRTR | CACAAAATCTCAAGAAGCTCGTCAT AGTAATGTGCACAGCCAACGAT |
| V-ATPase | PCATPRTFPCATPRTF | CCGTCGGTGATCCTGTTTTG ACCGGGACCCAATTCGA |
| 18S | PC18SRTFPC18SRTR | TCGGAAGGAACGCTTTTATTAGA ACGCCCGAAAGCGAAAC |
| GFP | GFP qPCR FGFP qPCR R | CCTGTCCTTTTACCAGACAACCA CACGCTTTTCGTTGGGATCT |
Primers for qRT-PCR studies were designed from target sequences using Primer Express 3.0 software and the best primers that did not produce secondary structures or primer dimers were selected. These primers targeted sequences flanking the input dsRNA sequences.
Statistical Analysis
Experimental data were analyzed to test the difference among treatments at a significance level of 0.05 and 0.01. Analysis of variance (ANOVA) was performed using Statistix 8.1. The homogenous groups are shown with similar letters based on least significant difference (LSD) values. Mortality was corrected using Abbot's formula [36].
Results
Sequence analysis
We successfully amplified and sequenced 507bp of CHS1, 1120bp of V-ATPase and 164bp of actin (ACTB) mRNAs from P. citri. To determine the sequence similarities of the three sequences from P. citri with those from other insect species, we performed BLAST analysis. The CHS1 sequence showed a maximum (87%) nucleotide identity with that of P. solenopsis (unpublished data) and a minimum (74%) with that of T. castaneum (Table S1 in Tables S1). V-ATPase sequence showed a maximum (83%) identity with that of P. solenopsis (unpublished data) and a minimum (76%) with that of A. albopictus (Table S2in Tables S1). BLAST search results of actin showed a maximum identity of 88% with that of P. solenopsis (unpublished data) and a minimum (76%) with that of Liposcelis entomophila (Table S3 in Tables S1). The BLAST search results validated the fidelity of the sequences amplified in our study.
RNAi induction by microinjection
In our experiments there was a significant (>40%) reduction of the target mRNAs in P. citri injected with effector dsRNAs as compared to insects injected with elution buffer (EB) or the control GFP dsRNA (Fig. 1A, B). Statistical analysis showed significant differences between samples injected with EB (control) and V-ATPase dsRNA, or those injected with GFP dsRNA and V-ATPase dsRNA showing an LSD value of 0.08 (P<0.01). Similarly there were significant differences between samples injected with EB and CHS1 dsRNA, or GFP dsRNA and CHS1 dsRNA showing an LSD value of 0.10 (P<0.01). There was no significant difference between samples injected with EB or GFP dsRNA.
Figure 1. Relative mRNA levels of CHS1 (A) and V-ATPase (B) following microinjection of specific dsRNAs into third instar P. citri.
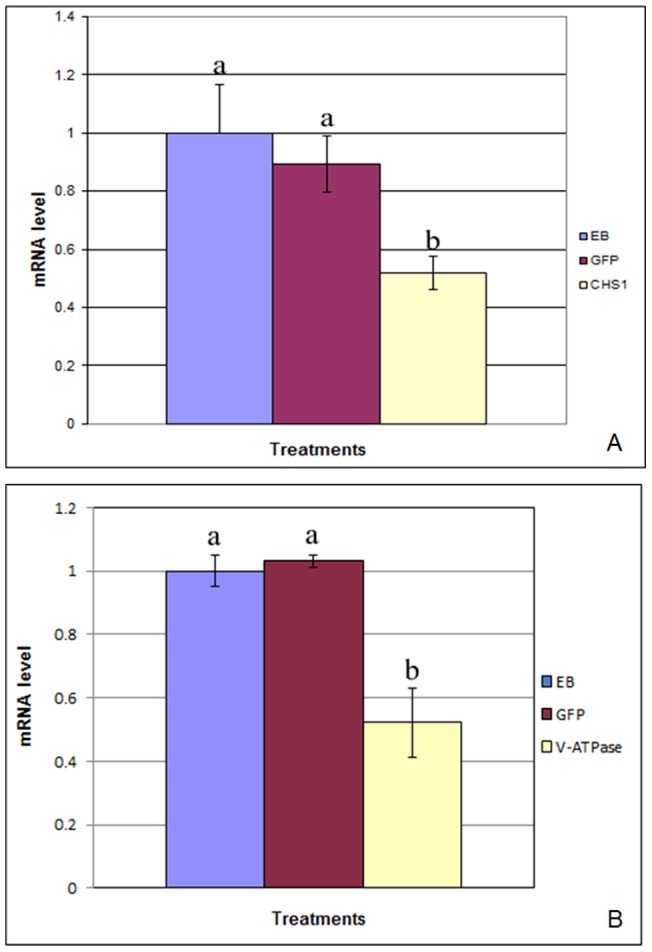
qRT-PCR results of CHS1 (A) and V-ATPase (B) mRNAs from P. citri following microinjection of elution buffer (EB), green fluorescent protein (GFP), CHS1 or V-ATPase dsRNAs. Total RNA was extracted 4 days post injection and used for cDNA synthesis. qRT-PCR was performed using primers described in Table 3. The P. citri 18S ribosomal RNA was used as an endogenous control in all experiments. Calculation of mRNA levels between control and treated groups was done by the comparative CT or 2–ΔΔCt method and analyzed by ANOVA using statistix 8.1 software. Numbers followed by the same letter are not different at p<0.01 (LSD 0.08). All experiments were performed twice and data were used collectively.
Recombinant TMV inoculated plants contain P. citri RNAs
In order to elucidate whether P. citri effector RNAs were produced in plants after inoculation using the recombinant TMVs, we first confirmed the presence of the actin, CHS1 and V-ATPase RNAs by RT-PCR. These plants tested positive by RT-PCR (Fig. 2A shows results for CHS1 and V-ATPase), by symptom development and by GFP fluorescence under UV light (for JL36-infected plants). These analyses identified recombinant TMVs containing actin RNA in the sense orientation (TMV-Actin), CHS1 RNA in sense (TMV-CHS1-S) and antisense (TMV-CHS1-AS) forms, and V-ATPase RNA in sense (TMV-V-ATPase-S) and antisense (TMV-V-ATPase-AS) forms. We further tested for siRNAs specific to the CHS1 RNA sequences by performing small RNA blot hybridization. RNAs from plants that were positive by RT-PCR also showed accumulation of the specific siRNAs (Fig. 2B).
Figure 2. Detection of P. citri CHS1 and V-ATPase mRNAs (A) and accumulation of RNAi-induced siRNAs (B) in N. benthamiana plants after TMV-CHS1 inoculation.
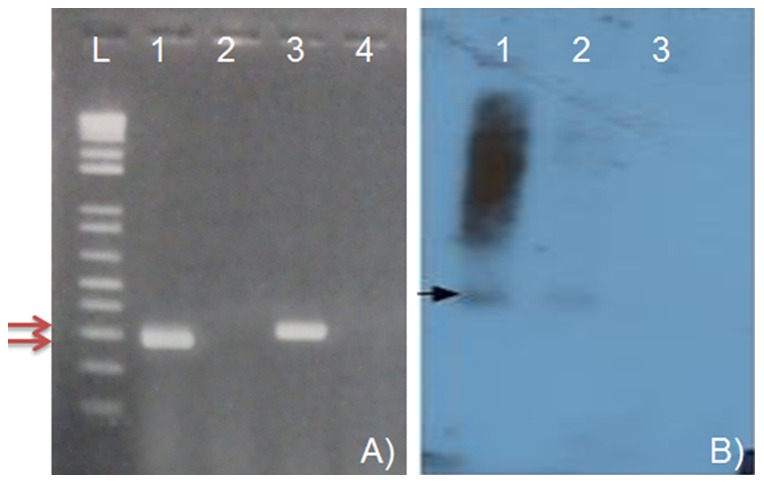
Plants were inoculated with recombinant TMV containing P. citri CHS1 (TMV-CHS1-S) and V-ATPase (TMV-V-ATPase-S). At 7 days post inoculation, total and small RNAs were isolated from infected plants and analyzed by RT-PCR and siRNA northern blot hybridization, respectively. A) One step RT-PCR was performed using total RNA as a template and CHS1 and V-ATPase primers and products analyzed on the gel. Lane L: 1Kb plus DNA ladder, Lane 1: CHS1 primers and RNA of TMV-CHS1-S inoculated N. benthamiana plants; Lane 2: CHS1 primers and RNA of TMV inoculated plant; Lane 3: V-ATPase primers and RNA of TMV-V-ATPase-S inoculated plant and Lane 4: V-ATPase primers and RNA of TMV inoculated plant. B) Small RNAs were separated by PAGE in 15% acrylamide, 8 M urea gels and processed for northern blot hybridization using 32P-UTP-labeled negative strand P.citri CHS1 transcript as a probe. Lane 1: TMV-CHS1-S inoculated plant; Lane 2: TMV-CHS1-AS iinoculated plant; Lane 3: TMV-inoculated plant.
Feeding on plants infected with recombinant TMVs induces RNAi in P. citri.
After 12 days of continuous feeding on plants infected with the recombinant TMVs, we collected P. citri for RNA extraction and qRT-PCR analyses. These analyses showed that there was ∼10% reduction of the P. citri CHS1 mRNA (Fig. 3A) for P. citri fed on plants infected with the TMV-CHS1-S and a 50% reduction for P. citri fed on plants infected with TMV-CHS1-AS. There was a significant difference among P. citri fed on control plants infected with TMV-GFP, and those infected with TMV-CHS1-AS with an LSD value of 0.04 (P<0.01). Similarly ∼30–40% reduction was seen for the V-ATPase mRNA (Fig. 3B) for P. citri fed on plants infected with both TMV-V-ATPase-S and TMV-V-ATPase-AS vs. those fed on plants infected with TMV-GFP, with an LSD value of 0.21 (P<0.01).
Figure 3. Relative levels of CHS1 (A) and V-ATPase (B) mRNAs in P. citri after feeding on N. benthamiana plants inoculated with recombinant TMV expressing P. citri CHS1 and V-ATPase fragments.
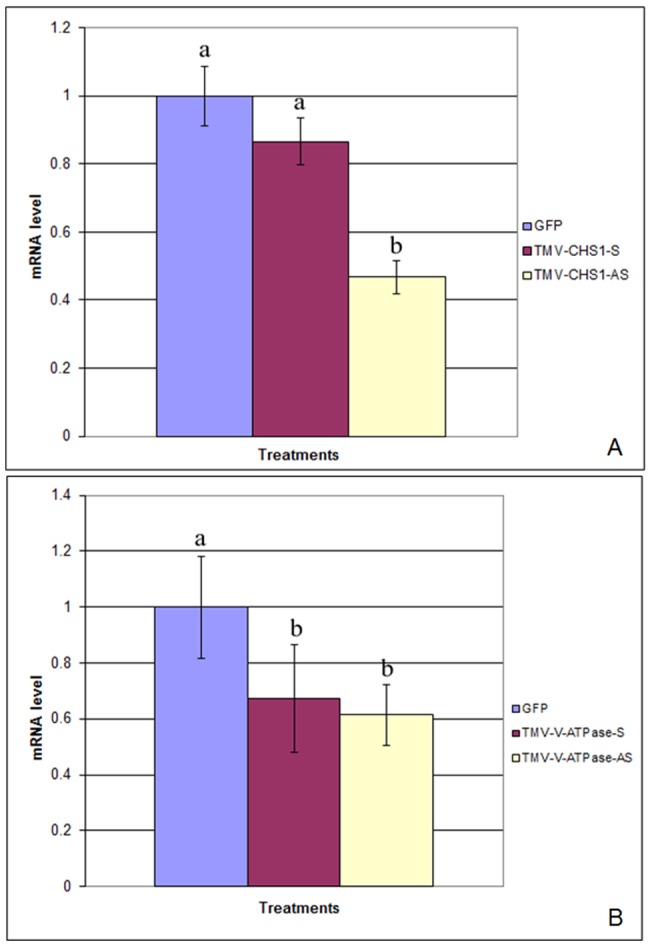
qRT-PCR results of CHS1 (A) and V-ATPase (B) mRNA levels from P. citri after feeding on N. benthamiana plants infected with recombinant TMVs. TMV-GFP (pJL24) was used as a control and sense and antisense inserts of P. citri CHS1/V-ATPase sequences were compared. Total RNA was extracted after 12 days feeding on plants and used for cDNA synthesis. qRT-PCR was performed using primers described in Table 3. The 18S ribosomal RNA was used as an endogenous control in all experiments. Calculation of mRNA levels between control and treated groups was carried out by the comparative CT or 2–ΔΔCt method and analyzed by ANOVA using statistix 8.1 software. Numbers with the same letter indicate homogenous groups at p<0.01 (LSD 0.04 and 0.21) for CHS1 and V-ATPase respectively. All experiments were performed twice and data were used collectively.
Reduction in P. citri actin mRNA correlates with reduced fecundity on recombinant TMV-Actin infected N. benthamiana plants
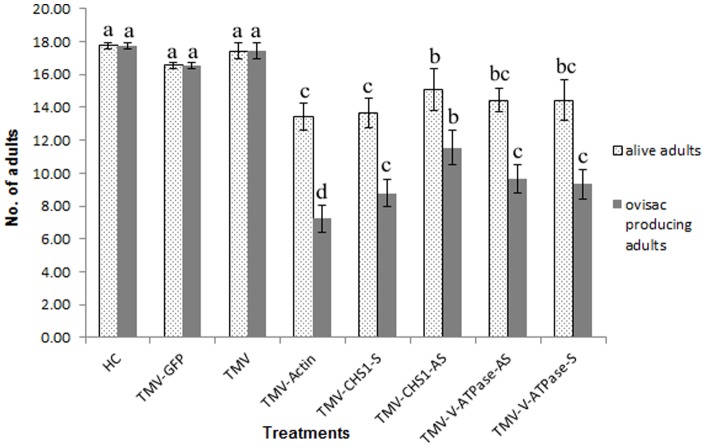
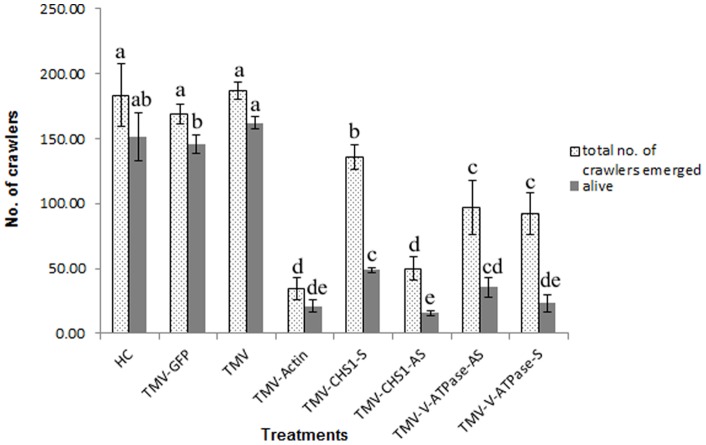
We observed that after continuous feeding on plants infected with recombinant TMV-Actin, there was no significant difference (P<0.05, LSD: 1.29) in survival of adult P. citri on all three control treatments: healthy plants, plants inoculated with TMV-GFP and plants inoculated with TMV only, showing mean survival of 17.78±0.19, 16.56±0.19 and 17.44±0.51, respectively. However, there was a significant difference between control groups and P. citri fed on TMV-Actin infected plants showing mean survival of 13.44±0.84. Similarly, there was a significant difference (P<0.05, LSD: 1.40) in the number of adults producing ovisacs for P. citri fed on healthy plants, plants inoculated with TMV-GFP, or on TMV-inoculated plants as compared to plants infected with TMV-Actin showing the mean number of adults producing ovisacs as 17.78±0.19, 16.56±0.19, 17.44±0.51 and 7.22±0.84 respectively (Figs. 4 & 5). Furthermore, crawlers emerging from ovisacs on TMV-Actin infected plants were greatly reduced (P<0.05, LSD: 25.69), with the mean number of emerging crawlers 34.44±8.39 (Fig. 4 & 6) as compared to the number of crawlers emerging from ovisacs on healthy control plants (183.33±24.04), on plants infected with TMV-GFP (168.89±7.70) and on plants that were infected with TMV only (186.67±6.67). There was no significant difference among all three control groups with respect to adult mortality, ovisac production, or crawler emergence and mortality in crawlers.
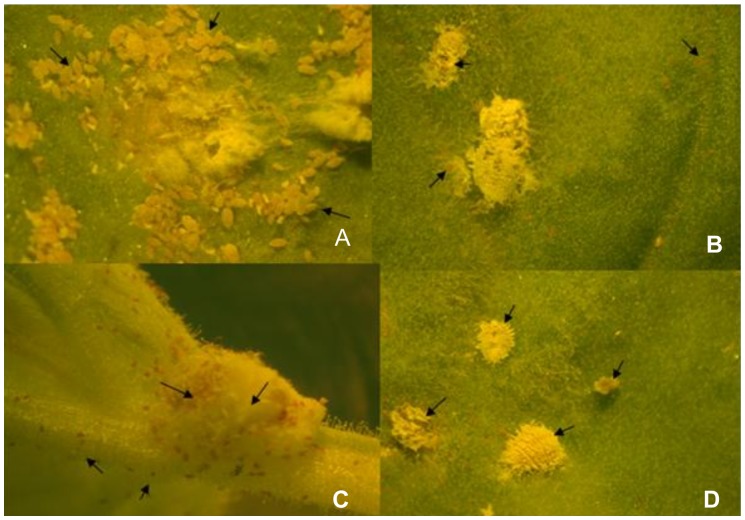
Figure 4. P. citri feeding on N. benthamiana plants inoculated with TMV-Actin.
Mealybug crawlers feeding on: A) N. benthamiana plants inoculated with TMV only (pJL36) showing healthy crawlers emerging while B, C and D show N. benthamiana plants inoculated with recombinant TMV (TMV-Actin) and show a very high mortality in crawlers and adults. Arrows in A indicate healthy crawlers; in B & C arrows indicate dead crawlers and D the arrows indicate dead adults.
Figure 5. Ovisac production and survival of adult P. citri fed on plants inoculated with different recombinant TMV constructs.
Number of surviving adult P. citri and ovisac production following 12 days feeding on plants inoculated with different recombinant TMV constructs. HC = healthy control N. benthamiana plants; TMV-GFP = TMV carrying GFP (TMV JL24); TMV = TMV with no insert (pJL36); TMV-Actin = part of the actin RNA inserted into TMV; TMV-CHS1-S and TMV-CHS1-AS = the chitin synthase 1 (CHS1) RNA fragment inserted into TMV in sense and antisense orientations, respectively; TMV-V-ATPase-S and TMV-V-ATPase-AS = the V-ATPase RNA fragment inserted into TMV in sense and antisense orientations, respectively. Numbers indicated with the same letter are homogenous groups at p<0.05.
Figure 6. Emergence of P. citri crawlers and their survival on plants inoculated with different recombinant TMV constructs.
Comparison of alive and total crawlers that emerged 18P. citri onto plants inoculated with the following viral constructs: HC = healthy control N. benthamiana plants; TMV-GFP = TMV carrying GFP (TMV JL24); TMV = TMV with no insert (pJL36); TMV-Actin = part of the actin RNA inserted into TMV; TMV-CHS1-S and TMV-CHS1-AS = the chitin synthase 1 (CHS1) RNA fragment inserted into TMV in sense and antisense orientations, respectively; TMV-V-ATPase-S and TMV-V-ATPase-AS = the V-ATPase RNA fragment inserted into TMV in sense and antisense orientations, respectively. Numbers indicated with the same letter are homogenous groups at p<0.05.
Decrease in mRNA level of CHS1 and V-ATPase correlates with decrease in fecundity and increased mortality of P. citri crawlers
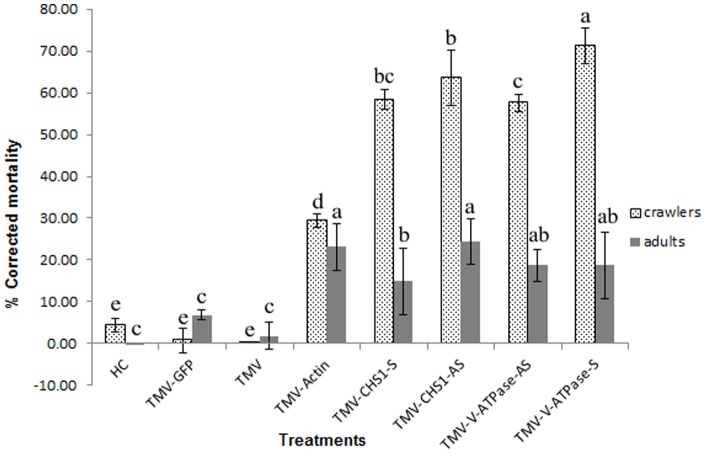
We observed that P. citri fed on plants infected with recombinant TMVs expressing the CHS1 and V-ATPase RNAs reached adult stage. The number of P. citri crawlers emerging from ovisacs and their survival when adults fed on plants infected with these recombinant TMVs were greatly reduced and significantly different from that seen for controls (Figs. 6 & 7). We also found that the mortality among P. citri placed on plants at the beginning of the bioassays were significantly lower than the mortality in the crawlers which emerged from the ovisacs of these parents (Fig. 8). Significant differences (P<0.05, LSD: 1.29) were also observed in survival of adult P. citri fed on plants infected with TMV-Actin, TMV-CHS1-S, TMV-CHS1-AS, TMV-V-ATPase-S, and TMV-V-ATPase-AS compared to the control groups. For P. citri fed on plants infected with TMV expressing CHS1 and V-ATPase RNA fragments there was a significant difference (P<0.05, LSD: 1.40) between the control and treated groups for ovisac production (TMV-CHS1-S 8.78±0.84, TMV-CHS1-AS 11.56±1.07, TMV-V-ATPase-S 9.33±0.88 and TMV-V-ATPase-AS 9.67±0.88). There was no significant difference between all three control groups, and similarly no significant difference between plants infected with TMV expressing sense and antisense V-ATPase sequences, but ovisac production was significantly lower in adults fed on plants infected with TMV-CHS1-S as compared to plants infected with TMV-CHS1-AS. A significant difference (P<0.05, LSD: 5.75) was seen between control groups (healthy plants 4.48±1.63, plants inoculated with TMV-GFP 0.82±2.94 and in plants inoculated with TMV only 0.00±0.00) and plants infected with the recombinant TMVs containing CHS1 and V-ATPase RNA sequences for mortality in crawlers showing mean corrected mortality of 58.41±2.39 and 63.66±6.63 for TMV-CHS1-S and TMV-CHS1-AS, respectively, and 71.22±4.22 and 57.67±2.07 for TMV-V-ATPase-S and TMV-V-ATPase-AS, respectively (Figs. 7 & 8).
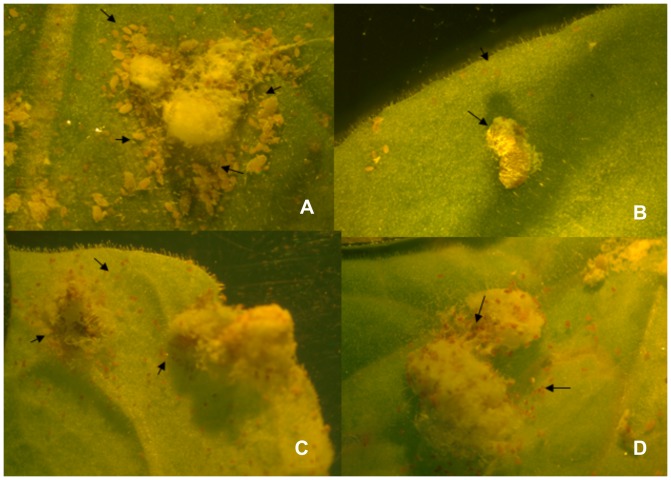
Figure 7. P. citri feeding on N. benthamiana plants inoculated with TMV-CHS1 and TMV-V-ATPase.
Mealybug crawlers feeding on: A) N. benthamiana plants infected with TMV (pJL36) show healthy crawlers emerging while B and C show N. benthamiana plants infected with TMV-CHS1-S where B shows abnormal ovisac formation and C shows a high mortality of crawlers. D shows an N. benthamiana plant infected with TMV-V-ATPase-S showing a high mortality in crawlers. Arrows in A indicate healthy crawlers, in B an abnormal ovisac and in C & D indicate dead crawlers.
Figure 8. Mortality in P. citri adults and crawlers on plants inoculated with different recombinant TMV constructs.
Comparison of corrected mortality in adult P. citri and crawlers following 18 days after release of insects onto plants inoculated with the following viral constructs: HC: healthy control N. benthamiana plant; TMV-GFP = TMV carrying GFP (TMV JL24); TMV = TMV with no insert (pJL36); TMV-Actin = part of the actin RNA inserted into TMV; TMV-CHS1-S and TMV-CHS1-AS = the chitin synthase 1 (CHS1) RNA fragment inserted into TMV in sense and antisense orientations, respectively; TMV-V-ATPase-S and TMV-V-ATPase-AS = the V-ATPase RNA fragment inserted into TMV in sense and antisense orientations, respectively. Numbers indicated with the same letter are homogenous groups at p<0.05.
Discussion
We first demonstrated RNAi effects in P. citri after injection of specific dsRNAs. This approach has been utilized by researchers [37], [38] on different insects for screening candidate targets for RNAi, but there is often high variability in terms of insect mortality due to the injection procedures, target RNAs and the target insect. Furthermore, evaluating RNAi effects for practical potential often does not reflect reality as injection is an artificial delivery system. For phloem-feeding hemipterans, such as P. citri, practical delivery is likely to be via the plants upon which the target insects feed. In order to achieve in planta evaluations of potential RNAi effectors, we used the recombinant virus-based vector, TMV. We hypothesized that plant viruses can be effective tools for targeting plant-feeding hemipterans by RNAi as viruses are both targets and powerful inducers of RNAi activity in plants [39]. Recently, Tobacco rattle virus (TRV) expression of antisense fragments from a chewing insect, M. sexta, in Nicotiana attenuata plants specifically silenced three midgut-expressed MsCYPs RNAs when larvae fed on these plants [32], and recombinant TMV was used to induce RNAi effects in Bactericera cockerelli on tomatillo plants [40]. Our data and that of Wuriyanghan and Falk [40] suggest that RNAi effectors can be present in the phloem in sufficient amounts to induce RNAi activity in phloem feeders, and other analyses have shown that siRNAs can be recovered from phloem sap [16].
We used the plant, N. benthamiana, due to its susceptibility to both the TMV and P. citri. We first confirmed by RT-PCR that the P. citri mRNA sequences were retained by recombinant TMV in infected plants and we used small RNA hybridization to demonstrate the presence of specific P. citri siRNAs in recombinant virus-infected plants. When P. citri were fed on N. benthamiana plants infected with recombinant TMV, we were able to detect specific RNAi effects including target mRNA reductions, P. citri mortality and reduced fecundity and hampered growth. Interestingly, the RNAi effects were much more prominent in the emerging nymphs/crawlers than in the adults which were initially placed to the TMV-inoculated plants. Wuriyanghan and Falk [40] also noted phenotypic effects on nymphs of the phloem-feeding B. cockerelli. Our combined data suggest that these effects may be due to the fact that the developing nymphs feed more voraciously than the older adults, hence can take up greater amounts of RNAi inducers as compared to adults, or they may be better RNAi targets.
Our qRT-PCR data showed significant reductions of the target mRNAs. Both dsRNA injection experiments and when P. citri were fed recombinant TMV-infected plants showed reductions in the corresponding mRNA target. By contrast, injection of GFP dsRNAs, or when P. citri fed on TMV-GFP-infected plants target mRNA reductions were not reduced. We cannot be sure that some non-target mRNAs were not also reduced (off target effects), but as the mRNAs evaluated here were not affected by GFP effectors and as we designed our sequences carefully using “SnapDragon” software (http://www.flyrnai.org/cgi-bin/RNAi_find_primers.pl), our data suggest that target mRNA reductions seen here were due to RNAi.
We also observed high mortality in adult mealybugs on plants infected with TMV-Actin and significant mortality (P<0.05) also on plants infected with TMV-CHS1 and TMV-V-ATPase. Interestingly we observed high mortality of 71.22±4.22 in the emerging P. citri crawlers on plants infected with TMV-V-ATPase. Crawler emergence and mortality were significantly different between samples fed on control group plants and TMV-V-ATPase infected plants. For P. citri fed on plants infected with TMV-CHS1, we found significant differences for plants infected with recombinant TMV having CHS1 inserts in the sense (TMV-CHS1-S) vs. antisense (TMV-CHS1-AS) orientations with respect to crawler emergence and mortality. Similar trends were also observed in qRT-PCR results where there was a significant difference in sense and antisense only for CHS1 constructs.
Previous studies have shown successful RNAi effects in insects through artificial feeding of siRNAs/dsRNAs, and through transient or stable transformation of plants expressing interfering RNAs [15]-[17], [19], [37], [41]–[43]. Our studies here provide another direction for rapid, yet functional in planta RNAi studies. TMV is easily manipulated, has a wide plant host range and gives fairly quick effects in plants. This approach provides a robust method to study the RNAi effects towards plant feeding insects, even phloem feeders, and the dramatic effects seen here by us particularly on nymphal/carwler mealybugs gives strong support for further efforts in RNAi-based insect control.
Supporting Information
Includes Table S1- Table S3.
(DOCX)
Acknowledgments
We acknowledge and appreciate Dr. Kristine Godfery for providing the P. citri culture and help in rearing P. citri. We also thank Dr. Jawwad A. Qureshi and Dr. Hada Wuriyanghan for their helpful suggestions in the research.
Funding Statement
The authors would like to thank Higher Education Commission of Pakistan for providing funding to Mr. Arif Muhammad Khan for conducting this research. This work also was supported in part by the University of California The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bivins JL, Deal JS (1973) Systemic insecticides for control of citrus mealybug in gardenias. Cal Agric 5.
- 2. Fayyaz-ur-Rehman M, Tariq MI, Aslam M, Khadija G, Iram A (2009) Inhibition studies of cellulolytic activities isolated from Planococcus citri. . Open Enz Inhib J 2: 8–11. [Google Scholar]
- 3. Hannon GJ (2002) RNA interference. Nature 418: 244–251. [DOI] [PubMed] [Google Scholar]
- 4. Kuttenkeuler D, Boutros M (2004) Genome-wide RNAi as a route to gene function in Drosophila . Brief Fun Genom Proteom 3: 168–176. [DOI] [PubMed] [Google Scholar]
- 5. Chen M, Du Q, Zhang H-Y, Wang X, Liang Z (2007) High-throughput screening using siRNA (RNAi) libraries. Expert Rev Mol Diag 7: 281–291. [DOI] [PubMed] [Google Scholar]
- 6. Price DRG, Gatehouse JA (2008) RNAi-mediated crop protection against insects. Tre Biotech 26: 393–400. [DOI] [PubMed] [Google Scholar]
- 7. Baum JA, Bogaert T, Clinton W, Heck GR, Feldmann P, et al. (2007) Control of coleopteran insect pests through RNA interference. Nat Biotech 25: 1322–1326. [DOI] [PubMed] [Google Scholar]
- 8. Gordon KH, Waterhouse PM (2007) RNAi for insect-proof plants. Nat Biotech 25: 1231–1232. [DOI] [PubMed] [Google Scholar]
- 9. Zhou X, Wheeler MM, Oi FM, Scharf ME (2008) RNA interference in the termite Reticulitermes flavipes through ingestion of double-stranded RNA. Insect Biochem Mol Bio 38: 805–815. [DOI] [PubMed] [Google Scholar]
- 10. Wuriyanghan H, Rosa C, Falk BW (2011) Oral delivery of double-stranded RNAs and siRNAs Induces RNAi effects in the potato/tomato psyllid, Bactericerca cockerelli . PLoS ONE 6: e27736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rosa C, Kamita SG, Falk BW (2012) RNA interference is induced in the glassy winged sharpshooter Homalodisca vitripennis by actin dsRNA. Pest Manag Sci 68: 995–1002. [DOI] [PubMed] [Google Scholar]
- 12. Rajagopal R, Sivakumar S, Agrawal N, Malhotra P, Bhatnagar RK (2002) Silencing of midgut aminopeptidase N of Spodoptera litura by double-stranded RNA establishes its role as Bacillus thuringiensis toxin receptor. J Biol Chem 277: 46849–46851. [DOI] [PubMed] [Google Scholar]
- 13. Bettencourt R, Terenius O, Faye I (2002) Hemolin gene silencing by ds-RNA injected into Cecropia pupae is lethal to next generation embryos. Insect Mol Bio 11: 267–271. [DOI] [PubMed] [Google Scholar]
- 14. Turner CT, Davy MW MacDiarmid RM Plummer KMBirch NP, et al. (2006) RNA interference in the light brown apple moth, Epiphyas postvittana (Walker) induced by double-stranded RNA feeding. Insect Mol Biol 15: 383–391. [DOI] [PubMed] [Google Scholar]
- 15. Zha W, Peng X, Chen R, Du B, Zhu L, et al. (2011) Knockdown of midgut genes by dsRNA-transgenic plant-mediated RNA interference in the hemipteran insect Nilaparvata lugens . PLoS ONE 6: e20504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pitino M, Coleman AD, Maffei ME, Ridout CJ, Hogenhout SA (2011) Silencing of aphid genes by dsRNA feeding from plants. PLoS ONE 6: e25709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mao YB, Cai WJ, Wang JW, Hong GJ, Tao XY, et al. (2007) Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat Biotech 25: 1307–1313. [DOI] [PubMed] [Google Scholar]
- 18. Whyard S, Singh AD, Wong S (2009) Ingested double-stranded RNAs can act as species-specific insecticides. Insect Biochem Mol Bio 39: 824–832. [DOI] [PubMed] [Google Scholar]
- 19. Mao YB, Tao XY, Xue XY, Wang LJ, Chen XY (2011) Cotton plants expressing CYP6AE14 double-stranded RNA show enhanced resistance to bollworms. Trans Res 20: 665–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Meyering-Vos M, Müller A (2007) RNA interference suggests sulfakinins as satiety effectors in the cricket Gryllus bimaculatus. J Insect Physio 53: 840–848. [DOI] [PubMed] [Google Scholar]
- 21. Swevers L, Liu J, Huvenne H, Smagghe G (2011) Search for limiting factors in the RNAi pathway in silkmoth tissues and the Bm5 cell line: the RNA-binding proteins R2D2 and translin. PLoS ONE 6: e20250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Terenius O, Papanicolaou A, Garbutt JS, Eleftherianos I, Huvenne H, et al. (2011) RNA interference in Lepidoptera: An overview of successful and unsuccessful studies and implications for experimental design. J Insect Physio 57: 231–245. [DOI] [PubMed] [Google Scholar]
- 23. Griebler M, Westerlund SA, Hoffmann KH, Meyering-Vos M (2008) RNA interference with the allatoregulating neuropeptide genes from the fall armyworm Spodoptera frugiperda and its effects on the JH titer in the hemolymph. J Insect Physio 54: 997–1007. [DOI] [PubMed] [Google Scholar]
- 24. Rodríguez-Cabrera L, Trujillo-Bacallao D, Borras-Hidalgo O, Wright DJ, Ayra-Pardo C (2010) RNAi-mediated knockdown of a Spodoptera frugiperda trypsin-like serine-protease gene reduces susceptibility to a Bacillus thuringiensis Cry1Ca1 protoxin. Env Microbio 12: 2894–2903. [DOI] [PubMed] [Google Scholar]
- 25. Kumar M, Gupta GP, Rajam MV (2009) Silencing of acetylcholinesterase gene of Helicoverpa armigera by siRNA affects larval growth and its life cycle. J Insect Physio 55: 273–278. [DOI] [PubMed] [Google Scholar]
- 26. Iga M, Smagghe G (2010) Identification and expression profile of Halloween genes involved in ecdysteroid biosynthesis in Spodoptera littoralis . Peptides 31: 456–467. [DOI] [PubMed] [Google Scholar]
- 27. Marcus JM (2005) Jumping genes and AFLP maps: transforming lepidopteran color pattern genetics. Evo & Dev 7: 108–114. [DOI] [PubMed] [Google Scholar]
- 28. Tomoyasu Y, Miller CS, Tomita S, Schoppmeier M, Grossmann Det al (2008) Exploring systemic RNA interference in insects: a genome-wide survey for RNAi genes in Tribolium . Geno Bio 9: R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Richards S, Gibbs RA, Weinstock GM, Brown SJ, Dennel R, et al. (2008) The genome of the model beetle and pest Tribolium castaneum. . Nature 452: 949–955. [DOI] [PubMed] [Google Scholar]
- 30. Jose AM, Hunter CP (2007) Transport of sequence-specific RNA interference information between cells. Ann Rev Gen 41: 305–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, et al. (2001) On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107: 465–476. [DOI] [PubMed] [Google Scholar]
- 32. Kumar P, Pandit SS, Baldwin IT (2012) Tobacco rattle virus vector: a rapid and transient means of silencing Manduca sexta genes by plant mediated RNA interference. PLoS One 7: e31347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lindbo J (2007) High-efficiency protein expression in plants from agroinfection-compatible Tobacco mosaic virus expression vectors. BMC Biotech 7: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bhaskar PB, Venkateshwaran M, Wu L, Ane J-M, Jiang J (2009) Agrobacterium-mediated transient gene expression and silencing: A rapid tool for functional gene assay in potato. PLoS ONE 4: e5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kim SW, Li Z, Moore PS, Monaghan AP, Chang Yet al (2010) A sensitive non-radioactive northern blot method to detect small RNAs. Nuc Aci Res 38: e98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Abbot WS (1925) A method of computing the effectiveness of an insecticide. J Eco Ento 18: 265–267. [Google Scholar]
- 37. Jaubert-Possamai S, LeTrionnaire G, Bonhomme J, Christophides G, Rispe C, et al. (2007) Gene knockdown by RNAi in the pea aphid Acyrthosiphon pisum . BMC Biotech 7: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yu N, Christiaens O, Liu J, Niu J, Cappelle K, et al. (2013) Delivery of dsRNA for RNAi in insects: an overview and future directions. Insect Sci 20: 4–14. [DOI] [PubMed] [Google Scholar]
- 39. Carrington JC, Kasschau KD, Johansen LK (2001) Activation and suppression of RNA silencing by plant viruses. Virology 281: 1–5. [DOI] [PubMed] [Google Scholar]
- 40. Wuriyanghan H, Falk BW (2013) RNA interference towards the potato psyllid, Bactericera cockerelli, is induced in plants infected with recombinant Tobacco mosaic virus (TMV). PLoS ONE 8(6): e66050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bucher G, Scholten J, Klingler M (2002) Parental RNAi in Tribolium (Coleoptera). Current Bio 12: R85–86. [DOI] [PubMed] [Google Scholar]
- 42. Araujo RN, Santos A, Pinto FS, Gontijo NF, Lehane MJ, et al. (2006) RNA interference of the salivary gland nitrophorin 2 in the Triatomine bug Rhodnius prolixus (Hemiptera: Reduviidae) by dsRNA ingestion or injection. Insect Biochem Mol Bio 36: 683–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mutti NS, Park Y, Reese JC, Reeck GR (2006) RNAi knockdown of a salivary transcript leading to lethality in the pea aphid, Acyrthosiphon pisum. . JInsect Sci 6: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Includes Table S1- Table S3.
(DOCX)