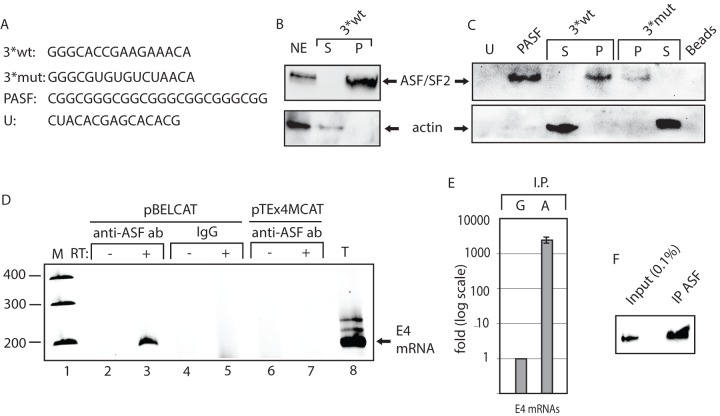
Figure 5. ASF/SF2 binds to site III in the splicing enhancer at HPV-16 SA3358.
(A). Sequences of the HPV-16 wild type (3*wt) and mutant (3*mut) enhancer site III sequence, four copies of an optimal ASF/SF2 binding site (PASF) serving as positive control and an unrelated RNA sequence termed “U”.The U sequence is adjacent to the ASF/SF2 site III in the HPV-16 genome. Biotinylated 2′-OMe-RNA forms of these sequences were used for RNA-protein pull-down experiments. (B) RNA-mediated pull down of ASF/SF2 from nuclear extracts protein usingbiotinylated, 2′-OMe-RNA sequences of wild type 3*wt RNA followed by Western blot analysis of RNA binding proteins in the pellet (P) or unbound proteins in the supernatant (S). (C) Western blot analysis of ASF/SF2 or actin after pull down of proteins using the various RNAs indicated in the figure. Pellet (P) represent the fraction of RNA bound proteins while unbound proteins are located in the supernatant (S). (D) RT-PCR on E4 mRNAs with primers 757s and E4A on cDNA synthesised from in vivo UV cross-linked, RNA-protein complexes immunoprecipitated with monospecific antibodies against ASF/SF2 (anti-ASF ab) or mouse IgG (IgG). Cells where either transfected with pBELCAT or the enhancer mutant pTEx4MCAT. RT-PCR was performed in the absence (−) or presence (+) of RT-enzyme for each sample. M, molecular weight markers; T, total cytoplasmic RNA. (E) Real time PCR of HPV-16 E4 mRNAs immunoprecipitated with mouse IgG (G) or anti-AFS/SF2 antibody (A) using primers 757s and E4A as described in Materials and Methods. (F) Western blot on ASF/SF2 immunoprecipitated by the murine anti-ASF/SF2 antibody revealed that approximately 2.3% of the ASF/SF2 proteins were immunoprecipitated.