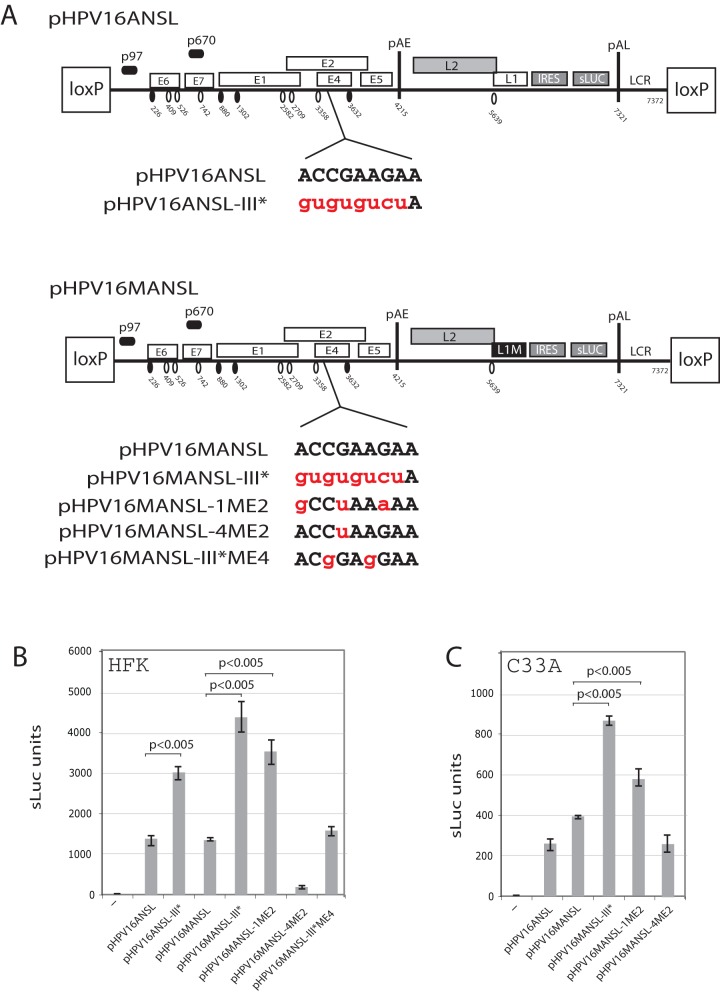
Figure 8. Nucleotide substitutions in ASF/SF2 binding site III in the splicing enhancer at SA3358 induced late gene expression from the full-length,episomalform of the HPV-16 genome.
(A). Schematic representation of the HPV-16 genomic plasmids pHPV16ANSL and pHPV16MANSL. The early and late viral promoters p97 and p670 are indicated. Numbers indicate nucleotide positions of 5′- (filled arrow heads) and 3′-splice sites (open arrow heads) or the early and late poly (A) sites pAE and pAL, respectively. L1M represents a previously described mutant HPV-16 L1 sequence in which a number of nucleotide substitutions inactivate splicing silencer elements downstream of late 3′-splice site SA5639. IRES, the poliovirus internal ribosome entry site sequence; sLuc, secreted luciferase gene; LCR, long control region. Sequences below plasmid maps represent wild type HPV-16 ASF/SF2 site III (capitals, black) and nucleotide substitutions (lower case, red) in the various plasmids indicated to the left. (B, C). Cell culture medium ofhuman primary keratinocytes collected at day 5 posttransfection with the various indicated plasmids was subjected to secreted-luciferase assay as described in Materials and Methods. Transfections were performed in the presence of pCAGGS-nlscre. Mean values and standard deviations of sLuc activity in the cell culture med of triplicate transfections are shown.