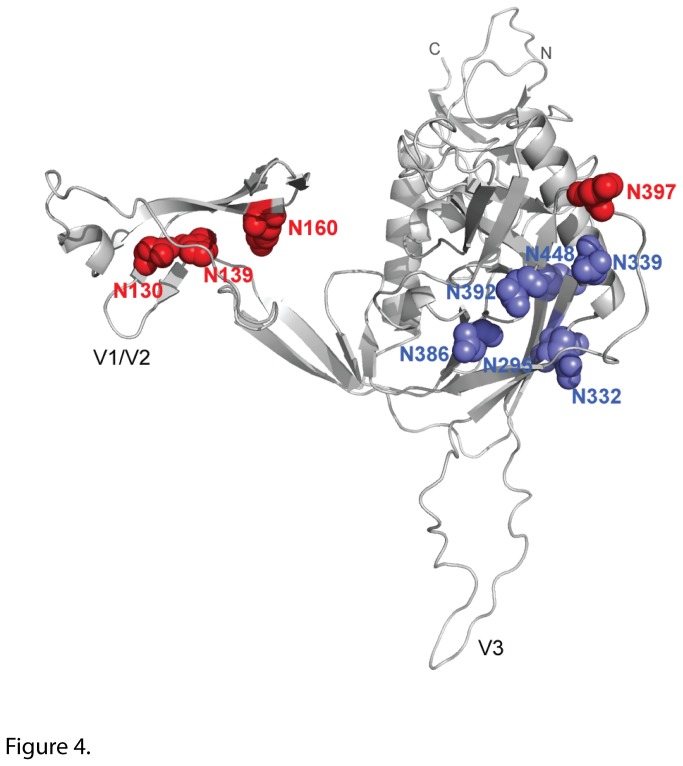
Figure 4. 3D model of PNGS of interest for 2G12 recognition.
The three-dimensional model of SF162 gp120 monomer (light grey) shows the location of the four PNGS of interest (red spheres) and the N-glycans (blue spheres) important for 2G12 binding. The model was built by homology modeling using the SWISS- MODEL server (http://swissmodel.expasy.org/) based on the structure of the HIV-1 JRFL gp120 core protein complexed with CD4 and the X5 antibody (PDB code 2B4C) [27]. The figure was generated with PyMOL Molecular Graphics System.