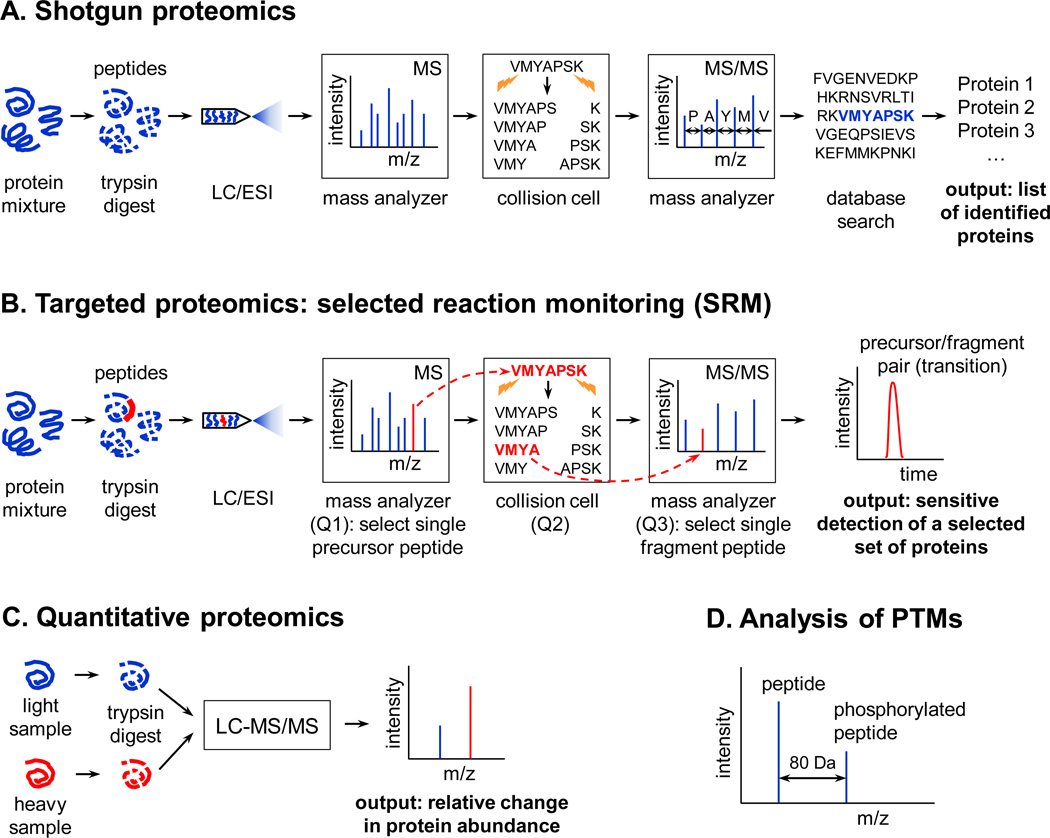
Figure 1.
Common mass spectrometry-based proteomics workflows. (A) Shotgun proteomics. The goal of shotgun proteomics experiments is to discover and identify as many proteins and their post-translational modifications (PTMs) as possible. An example fragmentation process in the collision cell is shown for a random peptide; this process is repeated for all detected peptides. (B) Targeted proteomics: selected reaction monitoring (SRM). The aim of targeted proteomics studies is to quantify a defined, limited set of proteins with high reproducibility and sensitivity. A single precursor peptide and its single fragment chosen for SRM are highlighted in red throughout the procedure. (C) Quantitative proteomics. The goal is to quantify differences in protein abundance between two or more samples, which is achieved by differential labeling of proteins or peptides with stable isotopes. This approach can be used both with shotgun and targeted proteomics methods. (D) Analysis of PTMs. Identification of PTMs is possible due to a mass shift that is characteristic for a particular modification. In the example shown, phosphorylation shifts the mass of a peptide by 80 Da. Abbreviations: LC, liquid chromatography; ESI, electrospray ionization. See text for more details.