Abstract
A 66-year-old woman heterozygous for a mutation in the ornithine transcarbamylase gene (Otc) participated in a phase I gene therapy trial for OTC deficiency. She received an adenovirus (Ad) vector expressing the functional OTC gene by intraportal perfusion. Fourteen years later she developed and subsequently died of hepatocellular carcinoma. A second subject, a 45-year-old woman, enrolled in the same trial presented with colon cancer 15 years later. We sought to investigate a possible association between the development of a tumor and prior adenoviral gene transfer in these two subjects. We developed and validated a sensitive nested polymerase chain reaction assay for recovering recombinant Ad sequences from host tissues. Using this method, we could not detect any Ad vector DNA in either tumor or normal tissue from the two patients. Our results are informative in ruling out the possibility that the adenoviral vector might have contributed to the development of cancer in those two subjects.
Zhong and colleagues use a nested PCR assay to examine whether adenoviral gene transfer led to the development of cancer in two clinical research subjects with ornithine transcarbamylase deficiency. Using this approach, they were unable to detect adenovirus vector genomes in normal or tumor tissues from the subjects more than a decade after vector infusion. These results suggest that adenoviral gene transfer was an unlikely contributor to the colon cancer or hepatocellular carcinoma observed in the two research subjects.
Introduction
Recombinant viral vectors are widely used in human gene therapy applications. Possible vector-related genotoxicity has gradually gained more attention. Viral vectors based on certain RNA viruses, such as murine retroviruses and lentiviruses, could have a particular predilection for integration into chromosomal DNA, and are therefore considered to carry a higher tumorigenesis potential. A gene therapy trial for the treatment of severe combined immunodeficiency using gamma retrovirus vector in fact demonstrated subsequent development of leukemia in several treated children and has re-emphasized such risks in viral gene therapy (Hacein-Bey-Abina et al., 2003; Kohn et al., 2003).
Although wild-type (wt) adenovirus (Ad) serotype 5 is capable of transforming cells in vitro via the E1a gene (van der Eb et al., 1977), natural infections of Ad in humans are not associated with cancer (Roy et al., 2009). In addition, the adenovirus is a double-stranded DNA virus that is not documented to have a propensity for integration into the host genome in vivo. The frequency of integration by E1-deleted replication-defective recombinant adenoviral vectors (rAd) detected in vivo ranged from 10−5 to 10−7 per transduced cell, no higher than plasmid DNA (Stephen et al., 2010). Moreover, rAd has been tested in a wide range of in vivo gene transfer applications for more than two decades. To date, there has been no reported evidence of an association between adenoviral gene therapy and transformation of host cells and tumor development. Therefore wild-type (wt)Ad and rAd are not considered to present a high risk for tumorigenesis (Wilson et al., 2012). However, a research subject enrolled in a gene therapy trial, who received the adenoviral vector expressing human ornithine transcarbamylase (H5.001CB.hOTC) at a dose 2×109 particles/kg via portal vein perfusion in April 1997 at the age of 52 years, was presented with hepatocellular carcinoma (HCC) 14 years later (Wilson et al., 2012). A second subject heterozygous for OTC deficiency received 2×1010 particles/kg at the age of 45 years in the same gene therapy trial and developed a colon malignancy 15 years later. Patients with urea cycle disorders have been found to be at an increased risk for liver dysfunction and hepatocellular damage, especially during hyperammonemic crises (Wilson et al., 2012). It has been speculated that these individuals also could have an increased risk for developing liver cancer (Stephen et al., 2010). On the other hand, no published studies have correlated either natural wtAd infections or rAd gene therapy with tumorigensis in humans. We hypothesized that the tumor formation in these two cases was not related to the rAd vector administration in the gene therapy trial more than a decade ago. The objective of our study was to formally test this hypothesis by molecular analysis of the tumor tissue DNA from the two study subjects for detection of vector sequences.
Results and Discussion
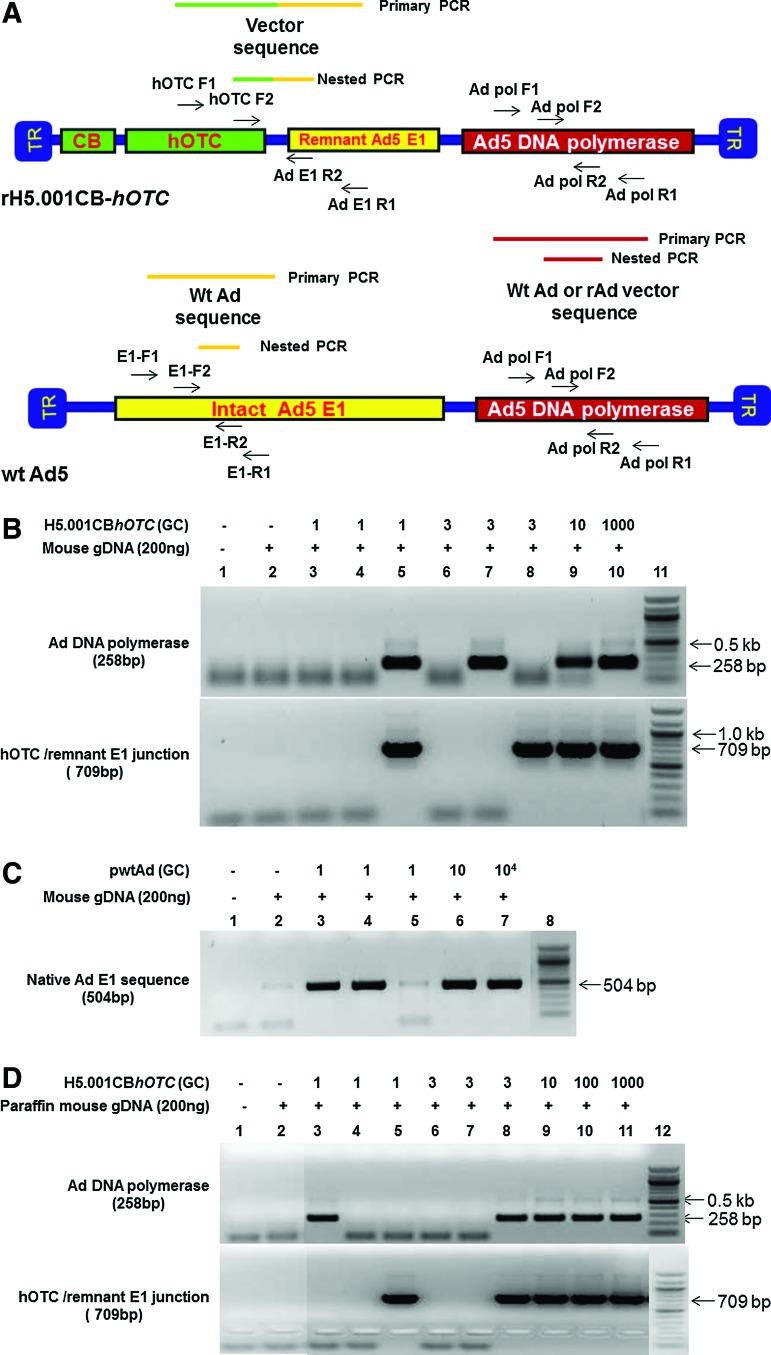
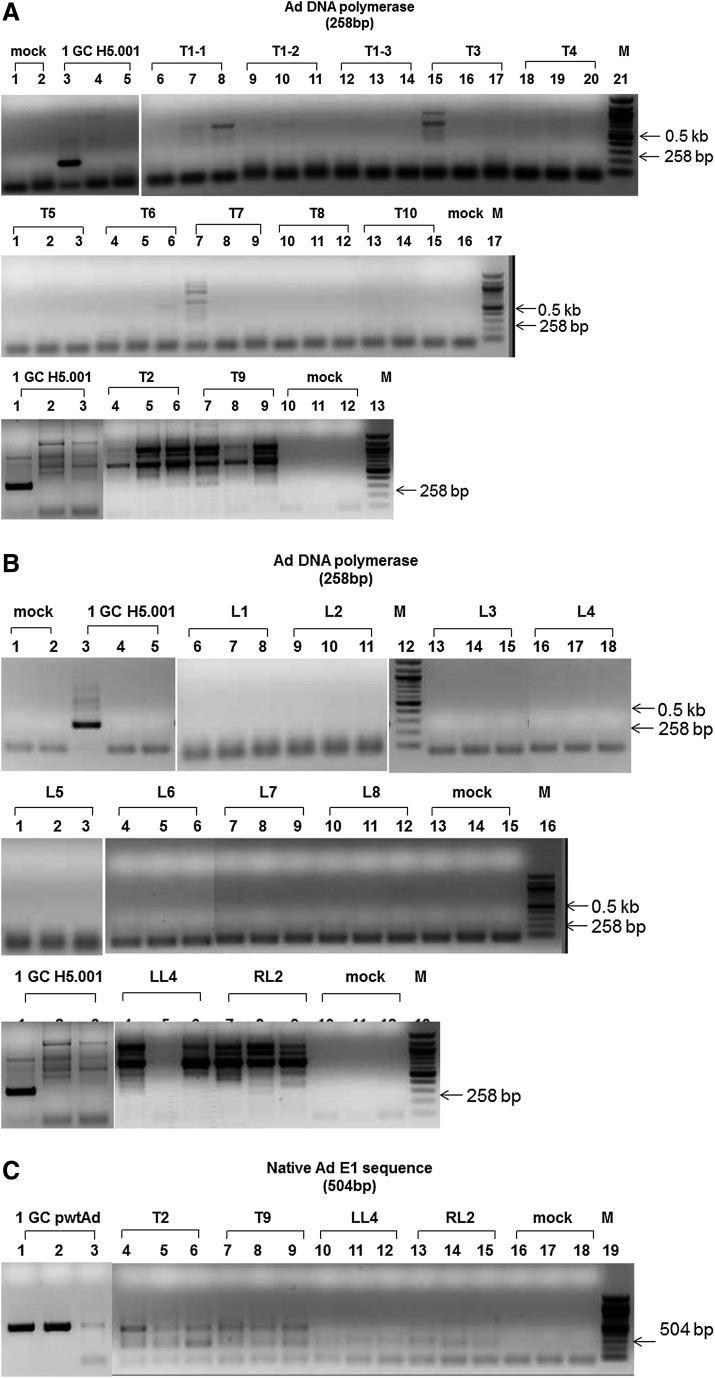
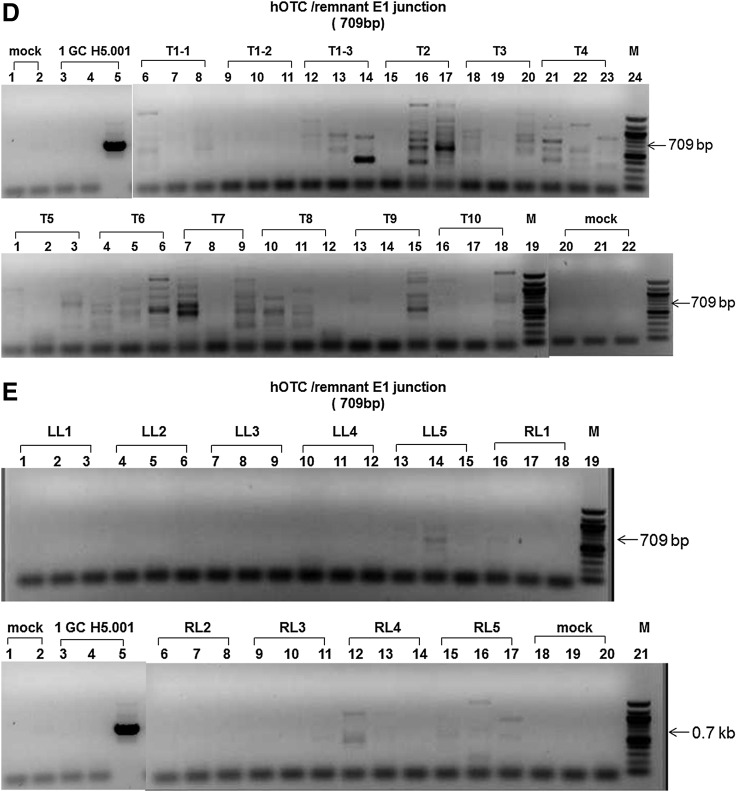
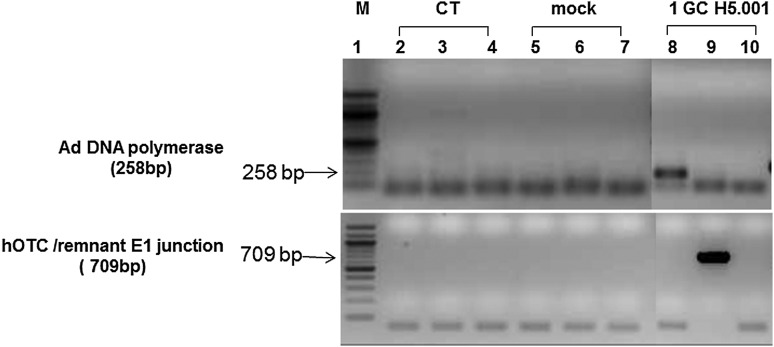
In this study, we developed, optimized, and validated a highly sensitive nested polymerase chain reaction (PCR) assay to recover the rAd or wtAd sequences in paraffin-embedded tissue from two Ad gene therapy study subjects who subsequently developed liver or colon malignancies. To this end, three sets of primer pairs were designed to amplify the native Ad5 DNA polymerase gene (Red in both rH5.001CB-hOTC vector and wtAd virus genomes), hOTC transgene/remnant E1 gene junction (Green and Yellow in rH5.001CB-hOTC vector only), and native E1 gene (Yellow in wtAd virus only) sequences (Fig. 1A and Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/humc). Our sequential PCR assay can detect as little as 1 genome copy of rAd or wtAd sequence spiked into 200 ng cellular DNA from frozen (Fig.1B and 1C, Table 1) or paraffin-embedded (Fig. 1D, Table 1) mouse liver tissue. Sequencing data confirmed that the PCR products were identical to the spiked rAd or wtAd sequences (data not shown). Using those optimized primers and PCR conditions, we then attempted to recover any possible rAd or wtAd sequences in the cellular DNA from normal liver, liver tumor, or colon tumor tissue isolated from those two patients. As summarized in Table 1, 10 normal liver samples and 12 liver tumor samples from patient 1 and one colon tumor sample from patient 2 were subjected to PCR for amplification of the native Ad5 DNA polymerase gene and the hOTC transgene/remnant E1 gene junction. Two normal liver samples and two liver tumor samples from patient 1 were also used for detecting the native E1 gene. No visible PCR products with the predicted sizes were amplified in any group (Table 1, Figs. 2 and 3). Some products with bands close to the predicted sizes were retrieved from the agarose gel and subjected to TOPO-cloning and sequencing. The sequencing data revealed that those products were nonspecific and did not contain any rAd or wtAd sequences (data not shown). Taken together, the data generated by these experiments indicated that neither normal tissue nor tumor tissue from the two patients contained any vector sequences introduced by the prior Ad vector gene therapy trial.
FIG. 1.
Primer design and assay validation for amplification of recombinant adenoviral vector (rAd) and wild-type adenoviral vector (wtAd) sequences in cellular DNA from normal mouse tissue by nested polymerase chain reaction (PCR). The location of the primary and nested PCR primers used for amplifications of the native Ad5 DNA polymerase gene (in both rH5.001CB-hOTC vector and wtAd virus), hOTC transgene/remnant E1 gene junction (in rH5.001CB-hOTC vector only) and native E1 gene (in wtAd virus only) sequences are schematically presented (A). To validate nested PCR using those primers, 200 ng each of cellular DNA from free (B, C) or paraffin-embedded (D) naive mouse liver tissue were spiked with different copy numbers (1–10,000 genome copies [GC]) of the isolated rH5.001CB-hOTC vector DNA (B, D) and wtAd plasmids (pH5wt) (C) and subjected to PCR. Nested PCR products were detected by 1.5% agarose gel electrophoresis and verified by TOPO-cloning followed by sequencing. The sensitivity of the PCR for amplifying target sequences was determined to one copy per reaction. OTC, ornithine transcarbamylase. Color images available online at www.liebertpub.com/hum
Table 1.
Detection of rAd and wtAd Sequences in Cellular DNA from Liver and Colon Tissue of OTC-Deficient Heterozygote Patients Who Received AdOTC Gene Therapy Trial More Than a Decade Ago
| |
|
|
|
Patient 1 (liver cancer) |
Patient 2 (colon cancer) |
|
|
|---|---|---|---|---|---|---|---|
| Sequence for amplification | Target for detection | Template for validation | PCR sensitivity (copy/reaction) | Normal tissue (10 samples) | Tumor tissue (12 samples) | Tumor tissue (1 sample) | Sequencing |
| Ad DNA polymerase gene | rAd and wtAd | rH5.001CB-hOTC vector DNA and wtAd5 plasmid DNA | 1 | — | — | — | Verified |
| hOTC transgene/remnant E1 gene junction | rAd | rH5.001CB-hOTC vector DNA | 1 | — | — | — | Verified |
| Native Ad E1 sequence | wtAd | wtAd5 plasmid DNA | 1 | —a | —a | — | Verified |
Two samples tested.
FIG. 2.
Detection of Ad sequences in cellular DNA from liver tissue of a patient who received AdOTC gene transfer. Using optimized primers and PCR conditions, 200 ng each of cellular DNA from liver tumor (A, C, D) and normal tissue (B, C, E) of an OTC deficiency heterozygote who received AdOTC gene transfer more than 10 years ago were subjected to nested PCR for amplifications of Ad5 DNA polymerase gene (A, B), hOTC transgene/remnant E1 gene junction (D, E), and native E1 gene (C) sequences. PCR amplification of Ad sequence in 200 ng each of cellular DNA from naive mouse liver tissue spiked with 1 GC of the isolated rH5.001CB-hOTC vector DNA (A, B, D, E) and wtAd plasmid (pH5wt) DNA (C) were used as positive controls. PCR products were detected by 1.5% agarose gel electrophoresis and verified by TOPO-cloning followed by sequencing. T, tumor; LL, left adjacent normal liver; RL, right adjacent normal liver; mock, no DNA template control; M, molecular weight ladder.
FIG. 3.
Detection of Ad sequences in cellular DNA from colon tissue of the second patient who participated in the AdOTC gene therapy trial. Using optimized primers and PCR conditions, 200 ng each of cellular DNA from paraffin-embedded colon tumor tissue of the second OTC deficiency patients who received AdOTC gene therapy more than 10 years ago were subjected to nested PCR for amplifications of the Ad5 DNA polymerase gene and hOTC transgene/remnant E1 gene junction sequences. PCR amplification of the Ad sequence in 200 ng each of paraffin-embedded cellular DNA from naive mouse liver tissue spiked with 1 GC of the isolated rH5.001CB-hOTC vector DNA were used as positive controls. PCR products were detected by 1.5% agarose gel electrophoresis and verified by TOPO-cloning followed by sequencing. CT, colon tumor; mock, no DNA template control; M, molecular weight ladder.
Conclusion
If the insertional activation by viral vector had triggered the tumorigenesis, the clonal expansion of the vector genome should have been observed in the tumor tissue. No adenovirus vector genome was detected in normal or tumor tissues from either subject after gene transfer more than a decade ago. Our data provide strong supporting evidence for our hypothesis that adenoviral gene transfer was an unlikely contributor to the HCC or colon cancer in the two patients. Instead, as discussed in our earlier report (Wilson et al., 2012), the liver dysfunction in urea cycle disorders may be implicated in HCC. In sum, we suggest that the tumor development in the two subjects is more likely to be attributable to the underlying OTC deficiency and its metabolic complications, and there is no direct evidence to support adenoviral gene transfer as a causative element. Nevertheless, one caveat of our present study is that the absence of vector sequences in tumor tissues does not eliminate the chances of clonal deletion of vector-positive cells during tumorigenesis, and the possible role of vector-expressed proteins influencing cellular events that may have caused transformation. Thus, further studies may be warranted to address these possibilities.
Supplementary Material
Acknowledgments
This research was supported by Public Health Service grant P01 HL59407-11 from the National Institutes of Health (to JMW and GG) and in part by a UMSS internal grant (to GG). It was also supported by the Rare Diseases Clinical Research Center on Urea Cycle Disorders (U54 HD061221).
Author Disclosure Statement
JMW is a consultant to ReGenX Holdings, and is a founder of, holds equity in, and receives a grant from affiliates of ReGenX Holdings; in addition, he is an inventor on patents licensed to various biopharmaceutical companies, including affiliates of ReGenX Holdings. The other authors report no competing financial interests.
References
- Hacein-Bey-Abina S. Von Kalle C. Schmidt M., et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- Kohn D.B. Sadelain M. Dunbar C., et al. American Society of Gene Therapy (ASGT) ad hoc subcommittee on retroviral-mediated gene transfer to hematopoietic stem cells. Mol. Ther. 2003;8:180–187. doi: 10.1016/s1525-0016(03)00212-0. [DOI] [PubMed] [Google Scholar]
- Roy S. Vandenberghe L.H. Kryazhimskiy S., et al. Isolation and characterization of adenoviruses persistently shed from the gastrointestinal tract of non-human primates. PLoS Pathog. 2009;5:e1000503. doi: 10.1371/journal.ppat.1000503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen S.L. Montini E. Sivanandam V.G., et al. Chromosomal integration of adenoviral vector DNA in vivo. J. Virol. 2010;84:9987–9994. doi: 10.1128/JVI.00751-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Eb A.J. Mulder C. Graham F.L. Houweling A. Transformation with specific fragments of adenovirus DNAs. I. Isolation of specific fragments with transforming activity of adenovirus 2 and 5 DNA. Gene. 1977;2:115–132. doi: 10.1016/0378-1119(77)90012-9. [DOI] [PubMed] [Google Scholar]
- Wilson J.M. Shchelochkov O.A. Gallagher R.C. Batshaw M.L. Hepatocellular carcinoma in a research subject with ornithine transcarbamylase deficiency. Mol. Genet. Metab. 2012;105:263–265. doi: 10.1016/j.ymgme.2011.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.