Figure 1.
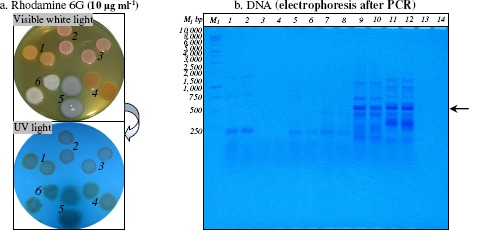
Rhodamine 6G accumulation in Gram-positive and Gram-negative bacterial cells (panel a.) and PCR amplification of HAE1 fragment using as template the DNA extracted from Gram-positive and Gram-negative bacteria (panel b.)
Panel a. After incubation at 28°C for 24 h, bacterial colonies was observed under visible white light and UV light; Mycobacterium sp. IBBPo1 (spots 1), Oerskovia sp. IBBPo2 (spots 2), Corynebacterium sp. IBBPo3 (spots 3), Chryseomonas sp. IBBPo7 (spots 4), Pseudomonas sp. IBBPo10 (spots 5), Burkholderia sp. IBBPo12 (spots 6).
Panel b. 1 kb DNA ladder, Promega (lane M1); Mycobacterium sp. IBBPo1 (lanes 1, 2), Oerskovia sp. IBBPo2 (lanes 3, 4), Corynebacterium sp. IBBPo3 (lanes 5, 6), Chryseomonas sp. IBBPo7 (lanes 7, 8), Pseudomonas sp. IBBPo10 (lanes 9, 10), Burkholderia sp. IBBPo12 (lanes 11, 12), negative control DNA (lanes 13, 14); anneling at 50.0°C (lanes 1, 3, 5, 7, 9, 11, 13), anneling at 52.0°C (lanes 2, 4, 6, 8, 10, 12, 14); the positions of the expected HAE1 fragment (550 bp) are indicated with arrow.
