Abstract
Proteolytic and/or lipolytic lactic acid bacteria (LAB) were isolated from visceral wastes of different fresh water fishes. LAB count was found to be highest in case of visceral wastes of Mrigal (5.88 log cfu/g) and lowest in that of tilapia (4.22 log cfu/g). Morphological, biochemical and molecular characterization of the selected LAB isolates were carried out. Two isolates FJ1 (E. faecalis NCIM5367) and LP3 (P. acidilactici NCIM5368) showed both proteolytic and lipolytic properties. All the six native isolates selected for characterization showed antagonistic properties against several human pathogens. All the native isolates were sensitive to antibiotics cephalothin and clindamycin; and, resistant to cotrimoxazole and vancomycin. Considering individually, P. acidilactici FM37, P. acidilactici MW2 and E. faecalis FD3 were sensitive to erythromycin. The two strains FJ1 (E. faecalis NCIM 5367) and LP3 (P. acidilactici NCIM 5368) that had both proteolytic and lipolytic properties have the potential for application in fermentative recovery of lipids and proteins from fish processing wastes.
Keywords: Lactic acid bacteria, fish waste, E. faecalis NCIM 5367, P. acidilactici NCIM 5368, lipids
INTRODUCTION
Current global fish production stands at about 141 million metric tonnes (MMT) and India generates considerable foreign exchange as well by exporting seafood in excess of 2.8 MMT apart from 3 MMT of freshwater fishes from the inland fisheries sector. At present, more than 76% of fish production is utilized globally for human consumption with the remaining going for miscellaneous purpose (13). Fish waste hitherto was not put to any alternative use and hence used to be dumped (16) leading to serious environmental problems. Both from marine and freshwater fish processing, the wastes generated include scales, skins, visceral mass (viscera, air bladder, gonads and other organs), head and fins. Unlike marine fish processing sector, fresh water fish processing is not well organized and hence presents a different level of waste generation and disposal. Among different types of waste generated, fish viscera alone contributes 15–25% of the total body weight. As per the recent estimate, global generation of fish industry waste stands in excess of 63 MMT and is known to be a good source of recoverable biomolecules (6). Researchers across the globe are focusing on methods to recover the biomolecules in order to reduce organic loading on the environment as well as decrease the pollution related problems (5, 26, 28).
Lactic acid fermentation is well known technology for recovery of biomolecules from various kinds of solid wastes generated from different industries based on animal processing including fish (2, 14, 26). Fish viscera are not only rich in different biomolecules but are also rich in beneficial lactic acid bacteria (LAB) with probiotics properties (4). Fermentation using LAB is an effective means of in situ acid generation in turn preserving the waste through ensilation (31) or allowing recovery of lipids/proteins (2). LAB have been used as probiotics due to their properties including antibacterial, immunodulation, control of intestinal homeostasis, resistance to gastric acidity, and bile acid resistance (34).
LAB is best known as starter culture (11) due to their versatile metabolic characteristics such as acidification activity, proteolytic activity and synthesis of metabolites like bacteriocin (10). It has been proved that microflora in the digestive tract of fish play an important role by producing antibacterial substances (35). Fermentation using LAB has a better impact than other conventional method like acid silaging as it is eco friendly, cost effective and also provides beneficial effects like antibacterial activity and antioxidative properties (30). Use of starter culture (LAB) makes the fermentation process more controllable as compared to use of in-situ LAB. Since LAB are reported to be very effective in recovery of biomolecules from fish industry waste (3, 12, 21), isolation of LAB from fish industry waste itself becomes all the more important. Against this background, the main objective of the present study was to isolate, characterize and identify native LAB from fish waste that have both lipolytic and/or proteolytic activity in order to explore them for simultaneous recovery of both lipids and proteins from fish industry waste.
MATERIALS AND METHODS
General
Fish viscera from freshly harvested commercial varieties of freshwater fishes [viz., Indian major carps (Rohu – Labeo rohita; Mrigal – Cirrhinus mrigala; and, Catla – Catla catla), common carp (Cyprinsu carpio) and tilapia (Oreochromis mossambicus)] were procured from local market and transported to laboratory under iced conditions. The pathogenic bacterial strains (viz., Bacillus cereus F4433, Escherichia coli MTCC118, Staphylococcus aureus FRI722, Salmonella paratyphi FB254, Salmonella typhi FB231, Salmonella itriditicus, Listeria monocytogenes Scott A, Yersinia enterocolitica MTCC859 and Micrococcus luteus) used in the study for antibacterial activity were from the institute culture collection (Food Microbiology Department, CFTRI, Mysore). Microbiological media were procured from M/s Hi-Media (Hi-Media, Mumbai, India), and polymerase chain reaction (PCR) reagents were from M/S Genei, Bangalore (India). All other chemicals were of analytical grade, unless otherwise mentioned. pH measurements in various samples was accomplished using pH metre (Cyberscan 1000, Eutech, Singapore) by directly immersing the combined glass calomel electrode into sample.
Isolation of proteolytic and lipolytic lactic acid bacteria (LAB)
Total LAB counts of the fish visceral waste samples were measured by plating serially diluted samples on MRS agar. Average of triplicate plates were taken to express the counts as log CFU (colony forming units) per gram of sample. LAB were isolated from viscera of different fresh water fishes and fish visceral waste (FVW) fermented naturally by the native LAB (NVW). FVW was obtained by mixing fish visceral waste with either jaggery (10% w/w) or glucose (10% w/w) as carbon source and 2% (w/w) sodium chloride to prevent spoilage microbes. LAB enriched in the NVW was isolated further. FVW and NVW were homogenized in physiological saline (1:10 w/v). Serially diluted of the sample were plated on MRS plates containing 1% (w/v) skimmed milk (pour plate method), incubated under anaerobic conditions at 37±1°C for 48h. The proteolytic LAB strains were identified by the presence of clear zone around the colonies. These colonies were picked up and further tested for their lipolytic activity by streaking them on to MRS plates containing 1% (v/v) of tributyrin as substrate. Lipolytic cultures were identified by clear zone around the colonies. Those cultures that were proteolytic and / or lipolytic were further characterized by different biochemical tests. These cultures were also subjected to evaluation of their antagonistic properties against Listeria monocytogenes Scott A and Micrococcus luteus.
Identification of LAB with potent lipolytic / proteolytic activity
Identification of LAB isolates from FVW was carried out as per the schemes outlined in the Bergey’s manual of Systematic Bacteriology (22). As the isolates Pediococcus acidilactici LP3, Pediococcus acidilactici MW2, Pediococcus acidilactici FM37, Enterococcus faecalis FJ1, Enterococcus faecalis FD3 and Pediococcus acidilactici FJ4 showed proteolytic and/or lioplytic activities along with antibacterial properties, molecular identification was undertaken only in case of these isolates. For molecular identification, the 16S ribosomal RNA (rRNA) gene was amplified using RNA primers-(5’TCATCTGTCCCACCTTGCGC3’) as forward and R (5’-GAGTTTGATCCTGGCTCAGG-3’) as reverse primer-as previously described (20). PCR was performed in the Thermocycler Gene AmpPCR system 9700 (Applied Biosystem, Perkin Elmer, Foster City, CA, USA) as per the standard protocol (29). Total cell lysate was prepared by harvesting the cell pellet and disrupting it in phosphate buffer saline (PBS pH 7.0; IX) containing 1% Triton X-100 (Sigma). The mixture was incubated in a boiling water bath for 15 min and subsequently snap-cooled in ice bath. Five micro litres of this lysate was used as template. Upon amplication, the PCR product was analysed by agarose (1%) gel electrophoresis. The PCR product was eluted from the gel, purified using Qiagen gel extraction kit (Qiagen, Hilden, Germany) and sent for nucleotide sequencing at M/s Vimta Laboratories, Hyderabad (India). Native isolates that were characterized biochemically were evaluated for antibacterial spectrum, antibiotic sensitivity and lipase activity. Two isolates that had higher proteolytic and/or lipolytic activity- i.e., E. faecalis FJ1 and P. acidilactici LP3 - were deposited in the national repository of industrial microorganisms at NCL, Pune (http://www.ncl-india.org) with the accession numbers NCIM 5367 and NCIM 5368, respectively.
Antibacterial spectrum of selected native LAB
Native LAB isolates that exhibited proteolytic and/or lipolytic activity and characterized by different biochemical tests were further tested for their antibacterial activity against different pathogens. The native LAB isolates were grown in MRS broth for 24h at 37°C under static condition. After incubation, the medium was centrifuged (Rotina 420R; Hettich, Germany) at 6000×g for 20 min to collect the supernatant which was designated as culture filtrate (CF). CF was neutralised with 0.1 N NaOH and was used for testing the antagonistic activity against different human pathogens, by agar well diffusion method (15). Selected pathogens were spread plated on MRS agar plates overlaid with soft Brain Heart Infusion (BHI) agar (0.8%) and allowed to grow for 4 – 6 hours. Further, an aliquot (50µl) of neutralized CF was added to the wells on these plates containing freshly grown pathogenic strains. The plates were then pre-incubated at 4˚C for 2–3h to allow for the test material to diffuse into agar and later they were incubated for a further 18 hours at 37˚C. Inhibition zones around the wells were measured after the incubation and activity graded based on the diameter of the zones (viz., zones with a diameter >6 and <10mm were graded as ‘+’, zones with >11 and <16mm diameter were graded as ‘++’; zones of more than 16mm diameter were graded ‘+++’; and, absence of zone was graded as ‘-’)
Antibiotic sensitivity test for selected native isolates
Native LAB isolates were evaluated for their antibiotic sensitivity using antibiotic sensitivity discs (M/s Hi-Media, Mumbai, India). MRS agar plates were overlaid with 8ml of MRS soft agar (0.8%) containing 50µl of freshly grown culture and the plates were incubated at 4˚C for the soft agar media to set. Antibiotic discs were placed on solid media and incubated at 37˚C for 24 h under anaerobic conditions. Inhibition zones were measured and results were interpreted as resistant (R), sensitive (S) or intermediate sensitive (IS) based on zones of inhibition as per manufacturer’s protocol.
Preparation of crude Enzyme extract and assaying the selected LAB for Lipase activity
Native LAB isolates from FVW were grown in MRS broth for 24 hours in an incubator shaker (150 rpm; Technico, India). Post incubation, the broth was centrifuged (Rotina 420R; Hettich, Germany) at 8000×g for 15 minutes to collect the culture filtrate which was designated as crude enzyme (CE) preparation. Lipase activity of CE was determined by estimating the free fatty acids (FFA) released upon hydrolysis of stabilized emulsion (SE) of vegetable oil by the CE as described by Borlongan (7). The stabilized emulsion was prepared by homogenizing 50 ml of sunflower oil with 50 ml of 2% bovine serum albumin and 3.5 ml of Tween 80. Assay was carried out by addition of 1ml of crude enzyme to 1ml of stabilized lipase substrate in 1.5 ml of 0.1M Tris–HCl buffer at pH 8.0. Mixture was incubated for 6 h at 37 °C, after which hydrolysis was stopped by addition of 3ml of 95% ethyl alcohol. The mixture was then titrated with 0.01N NaOH using 1% phenolphthalein in ethanol as indicator. For blank determination the same procedure was followed except that CE was introduced at the end of incubation period after addition of ethyl alcohol into the assay system. One unit of lipase activity was defined as the volume of 0.01N NaOH required to neutralize the FFA formed on hydrolysis of oil per milligram of protein in extract.
RESULTS
Quantitative and qualitative characteristics of LAB associated with fish waste
FVW and NVW had LAB counts in the range of 4.2 - 5.9 and 8 - 9 log cfu/g respectively (Table 1). LAB count was found to be highest in case of visceral wastes of Mrigal (5.88 log cfu/g) and lowest in that of tilapia (4.22 log cfu/g). The proteolytic and lipolytic properties of LAB isolates from FVW and/or NVW are shown in (Table 2).
Table 1.
Lactic acid bacteria (LAB) counts (log CFU/g) in visceral wastes of different freshwater fishes and isolates used for further characterization
| Sample | Count, log CFU | Codes of isolates picked up |
|---|---|---|
| Rohu (Labeo rohita) | 5.28 ± 0.25 | LP1, LP3, LP5 |
| Catla (Cirrhinus mrigala) | 5.12 ± 0.05 | FLB5, FS37, FS50, FS501 |
| Common carp (Catla catla) | 5.44 ± 0.17 | FM37,FM371, FM50, FM501 |
| Mrigal (Cyprinsu carpio) | 5.88 ± 0.10 | MW1, MW2, MW3, MW4 |
| Tilapia (Oreochromis mossambicus) | 4.22 ± 0.23 | TL |
| Naturally fermented sample | ||
| Jaggery | 8.45 ± 0.07 | FJ1,FJ4, |
| Dextrose | 8.08 ± 8.08 | FD1,FD2,FD3 |
Values indicate the average counts of triplicate plates of MRS agar CFU: colony forming units
Table 2.
Antibacterial-, proteolytic- and lipolytic activity of LAB isolated from freshwater fish viscera and naturally fermented fish viscera
| Fishes | Culture code | Identified* Genus | Antibacterial activity | Proteolytic activity | Lipase activity | |
|---|---|---|---|---|---|---|
| Listeria | Micrococcus | |||||
| Rohu | LP1 | Pediococcus | + | ++ | + | - |
| LP3* | Pediococcus | ++ | ++ | + | ++ | |
| LP5 | Pediococcus | - | ++ | + | - | |
| Catla | FLB5 | Enterococcus | ++ | ++ | + | - |
| FS37 | Enterococcus | ++ | ++ | - | - | |
| Common Carp | FM371 | Enterococcus | ++ | ++ | - | - |
| FM50 | Enterococcus | ++ | ++ | - | - | |
| FM501 | Pediococcus | - | ++ | - | - | |
| Mrigal | MW1 | Enterococcus | + | - | + | - |
| MW2* | Pediococcus | ++ | ++ | + | + | |
| MW4 | Enterococcus | ++ | ++ | + | + | |
| Tilapia | TL | Enterococcus | + | - | ++ | - |
| Naturally fermented fish visceral waste | ||||||
| Mixed waste | FJ1* | Enterococcus | ++ | ++ | ++ | ++ |
| FJ4* | Pediococcus | - | ++ | ++ | ++ | |
| FM37* | Pediococcus | ++ | - | - | + | |
| FD1 | Pediococcus | - | ++ | + | ++ | |
| FD2 | Enterococcus | - | ++ | + | + | |
| FD3* | Enterococcus | + | - | + | ++ | |
Identification based on different biochemical test (Grams staining, catalase test, Oxidase test, Growth at different pH, temperature and NaCl concentration, sugar fermentation test) +: moderate activity, ++: higher activity, -: negative
FVW and NVW samples were screened for LAB to obtain more than 100 discreet colonies. Of these, 30 isolates that exhibited both lipolytic and proteolytic activity (16 from FVW and 14 from NVW) were selected for further studies. 21 isolates (16 from FVW and 5 from NVW) out of the 30 considered exhibited higher lipolytic and proteolytic activities; and, these were coded and considered for choosing the best cultures (Table 1). Initial biochemical tests revealed the genus of these 21 isolates to be either Pediococcus or Enterococcus (Table 2). Among these 21 isolates, 6 isolates (LP3, MW2, FJ1, FJ4, FM37 and FD3) that exhibited antibacterial properties (against Listeria and / or Micrococcus) in addition to being lipolytic and proteolytic were chosen for further characterization and identification (Table 2).
The morphological, biochemical and physiological characteristics of these 6 isolates are presented in Table 3. The isolates LP3 and FM37 (both Pediococcus acidilactici) showed growth at 50°C, indicating their ability to tolerate relatively higher temperature; while, MW2 (Pediococcus acidilactici) showed growth in higher salt concentration (9%). All the isolates had no growth at 10 °C. All the isolates showed growth in the alkaline pH range (pH 7.5 and 8.5) and three isolates (FJ1, MW2, FD3) had no growth at pH 4.2. All 6 isolates viz., LP3, MW2, FJ1, FD3, FM37 and FJ4 were identified based on 16S rRNA homology (data not shown). These were identified as Pediococcus acidilactici LP3, P. acidilactici MW2, P. acidilactici FM37, Enterococcus faecalis FJ1, E. faecalis FD3 and P. acidilactici FJ4, respectively. From these tests, the isolates LP3, MW2, FM37 and FJ4 were identified as Pediococcus acidilactici whereas FJ1 and FD3 as Enterococcus faecalis (Table 3).
Table 3.
Physiological and biochemical growth characterstics of LAB isolates from fresh water fish visecra
| Biochemical Assay | LP3 | FJ1 | MW2 | FM37 | FD3 | FJ4 | |
|---|---|---|---|---|---|---|---|
| Morphology | cocci | cocci | cocci | cocci | Cocci | cocci | |
| Gram Staining | + | + | + | + | + | + | |
| Catalase / Oxidase | - / + | - / + | - / + | - / + | - / + | - / + | |
| Growth temperature (°C) | 10 | - | - | - | - | - | - |
| 30 | + | + | + | + | + | + | |
| 50 | + | - | - | + | - | - | |
| Growth at NaCl (%) | 3.0 | + | + | + | + | + | + |
| 6.5 | + | + | + | - | + | + | |
| 9.0 | - | - | + | - | - | - | |
| Growth at pH | 4.2 | + | - | - | + | - | + |
| 7.5 | + | + | + | + | + | + | |
| 8.5 | + | + | + | + | + | + | |
| Casein utilization | + | + | + | + | + | + | |
| Molecular identification | P. acidilactici NCIM5368 | E. faecalis NCIM 5367 | P.acidilactici | P. acidilactici | E. faecalis | P. acidilactici | |
Antibacterial activity and antibiotic sensitivity pattern of LAB isolates
The antagonistic properties 6 different LAB isolates selected for further studies is presented in Table 4. The native LAB isolates from FVW, generally, indicated antagonistic properties against different bacterial pathogens employed in the study (Table 4) pointing to their broad spectrum of activity against these pathogens. However, L. monocytogenes Scott A was not inhibited by P. acidilactici FJ4, Y. enterocolitica MTCC859 was not inhibited by P. acidilactici: FM37 and B. cereus not inhibited by P. acidilactici LP3.
Table 4.
Antibacterial spectrum of four selected native isolates against different pathogens
| Pathogens | Isolates | |||||
|---|---|---|---|---|---|---|
| LP3 | FJ1 | MW2 | FM37 | FD3 | FJ4 | |
| L. monocytogenes Scott A | ++ | ++ | ++ | ++ | + | - |
| M. luteus | ++ | ++ | ++ | ++ | - | ++ |
| S .typhi FB231 | + | ++ | ++ | ++ | + | ++ |
| S.itriditicus FB256 | ++ | +++ | +++ | +++ | ++ | +++ |
| S. paratyphi FB254 | ++ | ++ | + | ++ | ++ | + |
| Y. enterocolitica TCC859 | + | + | ++ | - | + | + |
| E. coli MTCC118 | ++ | +++ | ++ | +++ | ++ | +++ |
| S. aureus FR1722 | ++ | ++ | ++ | ++ | ++ | ++ |
| B. cereus F4433 | - | ++ | ++ | ++ | + | +++ |
LP3: Pediococcus acidilactici NCIM 5368, FJ1: Enterococcus faecalis NCIM 5367, MW2: Pediococcus acidilactici, FM37: Pediococcus acidilactici, FD3: Enterococcus faecalis, FJ4: Pediococcus acidilactici
( ‘-‘absence of zones, ‘+’ 10mm diameter zones, ‘++’ 16mm diameter zones ‘+++’ More than16mm diameter )
Further, antibiotic sensitivity assay revealed that the native LAB isolate’s sensitivity towards different antibiotics (Table 5). All the native isolates were sensitive to antibiotics cephalothin and clindamycin; and, resistant to cotrimoxazole and vancomycin. Considering individually, P. acidilactici FM37, P. acidilactici MW2 and E. faecalis FD3 were sensitive to erythromycin. Apart from this, the LAB isolates had varying degree of sensitivity towards different antibiotics (Table 5).
Table 5.
Antibiotic sensitivity test for native LAB isolated from fresh water fish viscera
| Antibiotics | Concentration (µg) | Isolates | |||||
|---|---|---|---|---|---|---|---|
| LP3 | FJ1 | MW2 | FM37 | FD3 | FJ4 | ||
| Cephalothin (Ch) | 30 | S | S | S | S | S | S |
| Clindamycin (cd) | 2 | S | S | S | S | S | S |
| CoTrimoxazole (co) | 25 | R | R | R | R | R | R |
| Erythromycin (E) | 15 | R | R | S | S | S | R |
| Gentamycin(G) | 10 | S | S | IM | R | IM | R |
| Ofloxacin (OF) | 1 | R | S | R | R | R | R |
| Pencillin-G (P) | 10 units | IM | IM | IM | R | R | R |
| Vancomycin (Van) | 30 | R | R | R | R | R | R |
S: sensitive, R: resistant and IM: Intermediate
LP3: Pediococcus acidilactici NCIM 5368, FJ1: Enterococcus faecalis NCIM 5367, MW2: Pediococcus acidilactici FM37: Pediococcus acidilactici, FD3: Enterococcus faecalis, FJ4: Pediococcus acidilactici
All the 6 isolates were grown with or without a lipid substrate and in-vitro lipase assay was carried out using CE and the results are presented in Figure 1. The results clearly indicated that all the isolates are capable of producing extracellular lipase; and, two isolates LP3 (P. acidilactici) and FJ1 (E. faecalis) exhibited higher lipolytic activity. These characteristics pointed to their potential for application in reclamation of different biomolecules, especially lipids and proteins from fish processing wastes. Hence, from among the 6 identified LAB isolates, two isolates P. acidilactici LP3 and E. faecalis FJ1 which exhibited higher lipolytic (Figure 1; Table 2) and proteolytic activity in addition to being antagonistic towards Listeria and/or Micrococcus (Table 2) were deposited in the national repository of industrial microorganisms at National Chemical Laboratory (NCL), Pune (http://www.ncl-india.org) with the accession numbers NCIM 5367(E. faecalis FJ1) and NCIM 5368 (P. acidilactici LP3).
Figure 1.
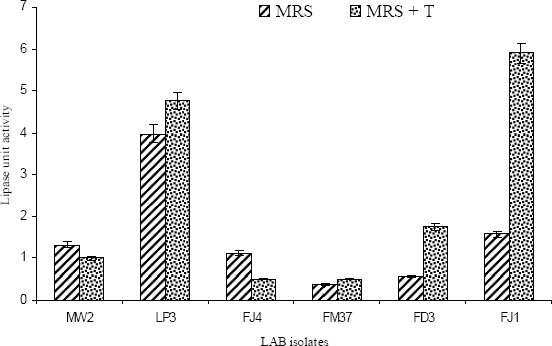
Lipolytic activity of LAB isolated from fresh fish viscera (n=3). (MRS+T = MRS with 1% Tributyrin as lipid substrate)
DISCUSSION
LAB having proteolytic, lipolytic and antibacterial properties against different pathogens were isolated from viscera of different fresh water fishes. The LAB isolated from FVW were found to be either Enterococcus or Pediococcus. However, several researchers have reported Lactobacillus to be the major LAB present in fish viscera (8) or meat (25). LAB composition in fish intestine is affected by seasonal changes, water temperature changes (19) which possibly explains the differences in observations about LAB. Also, these factors coupled with difference in LAB load in the wastes, make natural fermentation quite ineffective for oil or protein recovery from fish industry wastes. In addition, several researchers have reported the usefulness of LAB isolated from the same source, in comparison to LAB from other sources, as a better starter culture for fermentation of similar or same products (24, 2). Hence, use of single species of LAB as a starter culture make the fermentative hydrolysis more controllable, apart from hastening it, thereby making it reproducible. Presence of proteolytic and lipolytic LAB has been reported in viscera of fresh water fishes and marine shellfish waste (32, 33).
Fermentation with LAB has resulted in liquor portion rich in hydrolysed proteins which possess antioxidant activities as reported in case fermented shrimp waste (30) and tannery fleshings (2). Addition of LAB to ferment fish waste results in the domination of other microorganisms present in waste by the added LAB followed by reduction of pH in the fermented mass (18, 12) and this explains the preservative properties of LAB fermentation. In addition, mild organic acids produced also contribute towards hydrolysing proteins into low molecular peptides. In all, proteolytic LAB presumably hydrolyzes the protein-lipid complex, after which protein released probably gets further hydrolysed into smaller peptides due to the proteolytic action of LAB as well as the lactic acid produced by them. These hydrolysed proteins presumably exhibit biofunctional activities like antioxidant properties. Apart from lipolytic and proteolytic properties bacteriocin produced by these LAB can also provide antibacterial properties to the hydrolysate. Among all the native isolates Enterococcus faecalis NCIM5367 (FJ1) and Pediococcus acidilactici FJ4, isolated from naturally fermented fish waste had higher proteolytic and lipolytic properties compared to other isolates. The selected LAB isolated from NVW showed higher lipolytic properties compared to isolates from FVW of different fresh water fishes.
In our initial experiment for screening of LAB producing antibacterial compound Listeria monocytogenes Scott A and Micrococcus luteus were used. Six native isolates showing better proteolytic, lipolytic and antibacterial properties were further characterized and identified. Presence of LAB having antibacterial properties has been reported by several researchers (17, 27). The native LAB isolates from FVW or NVW, generally, indicated antagonistic properties against different bacterial pathogens employed in the study. Several authors have reported the antibacterial properties of LAB. For instance, Campos et al (9) isolated LAB strains from turbot muscle that exhibited inhibitory activity against Listeria monocytogenes and Staphylococcus aureus; while, Kalaou et al. (23) studied the activity of LAB on some Gram positive and negative pathogenic bacteria such as, Escherichia coli, Pseudomonas aeroginosa, Klebsiella pneumoniae, Staphylococcus aureus and Bacillus cereus and the inhibition zones were in the range of 1.4 to 2.8 cm. It has proved that Bacillus sp isolated from the fish gut have potential antagonistic effect against the fish pathogen Aeromonas hydrophila (35). In our study, the selected LAB isolates exhibited antagonistic properties against different human as well as fish pathogens. The antibacterial activity exhibited by the liquor portion rich in hydrolysed portion could possibly be due to the presence of antimicrobial peptides (bacteriocin) produced by the native LAB. This fraction has a potential to be used as an ingredient in livestock or aquaculture feeds. This can be used as a probiotic; and, use of probiotic as feed additives is preferred over that of antibiotics as they do not exhibit any of the undesirable effects associated with the use of antibiotics viz, toxicity, allergy, residues in food, bacterial drug resistance (1).
CONCLUSION
Out of the LAB isolated from FVW and/or NVW, two isolates P. acidilactici LP3 and E. faecalis FJ1 exhibited higher lipolytic and proteolytic activity in addition to being antagonistic towards bacterial pathogens. These were deposited in the national repository of industrial microorganisms with the accession numbers NCIM 5367(E. faecalis FJ1) and NCIM 5368 (P. acidilactici LP3). The production of lipase as a metabolite indicates the potential of these cultures for application in simultaneous recovery of both proteins and lipids apart from in-situ enrichment of PUFA in the recovered fish oil. Further studies are required to evaluate the effect of these native isolates on in-situ enrichment of PUFA in fish oil recovered from fish processing wastes.
ACKNOWLEDGEMENTS
Authors thank Department of Biotechnology (DBT), Govt. of India for partial funding of this work through Grant ##BT/PR 9474/AAQ/03/345/2007. Authors place on record their thanks to Dr V Prakash, Director, CFTRI for encouragement and permission to publish the work.
REFERENCES
- 1.Ajitha S. M., Sridhar N., Singh I.S.B., Varghese V. Probiotic effects of lactic acid bacteria against Vibirio alginolyticus in Penaeus (Fenneropenaeus) indicus (H. Milne Edwards) Asian Fisheries Sci. 2004;17:71–80. [Google Scholar]
- 2.Amit K.R., Bhaskar N., Halami P.M., Indirani K., Suresh P.V., Mahendrakar N.S. Characterization and application of a native lactic acid bacterium isolated from tannery fleshings for the fermentative bioconversion of tannery fleshing. Appl. Microbiol. Biotechnol. 2009;83:757–766. doi: 10.1007/s00253-009-1970-3. [DOI] [PubMed] [Google Scholar]
- 3.Amit K.R., Swapna H.C., Bhaskar N., Halami P.M., Sachindra N.M. Effect of fermentation ensilaging on recovery of oil from fresh water fish viscera. Enz. Micro. Technol. 2010;46:9–13. [Google Scholar]
- 4.Balcazar J.L., Venderll D., de Blas I., Ruiz-Zarzuela I., Muzquiz J.L., Girones O. Characterization of probiotic properties of lactic acid bacteria isolated from intestinal microbiota of fish. Aquaculture. 2008;278:188–219. [Google Scholar]
- 5.Bhaskar N., Benila T., Radha C., Lalitha R.G. Optimization of enzymatic hydrolysis of visceral waste proteins of Catla (Catla catla) for preparing protein hydrolysate using a commercial protease. Biores. Technol. 2008;99:335–343. doi: 10.1016/j.biortech.2006.12.015. [DOI] [PubMed] [Google Scholar]
- 6.Bhaskar N., Sachindra N.M., Suresh P.V., Mahendrakar N.S. Microbial Reclamation of Fish Industry By-products, in Press. In: Montet D., Ray R.C., editors. Aquaculture microbiology and biotechnology. vol. 2. Enfield, NH, USA: Science Publ Inc.; 2010. [Google Scholar]
- 7.Borlongan I.G. Studies on digestive lipases of milkfish. Aquaculture. 1990;89:315–325. [Google Scholar]
- 8.Cai Y., Suyanandana P., Saman P., Benno Y. Classfication and characterization of lactic acid bacteria isolated from the intestine of common crap and fresh water prawns. J. Gen. Appl. Microbiol. 1999;45:177–184. doi: 10.2323/jgam.45.177. [DOI] [PubMed] [Google Scholar]
- 9.Campos C.A., Rodríguez O., Calo-Mata P., Prado M., Barros Velázquez J. Preliminary characterization of bacteriocins from Lactococcus lactis, Enterococcus faecium and Enterococcus mundtii strains isolated from turbot (Psetta maxima) Food Res. Int. 2006;39:356–364. [Google Scholar]
- 10.Cheriguene A., Choguani F., Benslotane A. Identification and Characterization of Lactic Acid Bacteria Isolated from Algeris Goat’s Milk. Pakistan J. Biological Sci. 2006;7:1242–1249. [Google Scholar]
- 11.Cintas M., Herranz C., Hernández P.E., Casaus M.P., Nes L.F. Review: Bacteriocins of lactic acid bacteria. Food Sci. Tech Int. 2001;7:281–305. [Google Scholar]
- 12.Ennouali M., Elmoualdi L., Labioui H., Ouhsine M., Elyachioui M. Biotransformation of the fish waste by fermentation. African J. Biotechnol. 2006;5:1733–1737. [Google Scholar]
- 13.FAO . 2010. [last accessed on July 21, 2010]. Year Book of Fishery Statistics—Latest summary of tables. ftp://ftp.fao.org/FI/STAT/SUMM_TAB.HTM. [Google Scholar]
- 14.Gao M.T., Hirata M., Toorisaka E., Hano T. Acid –hydrolysis of fish wastes for lactic acid bacteria fermentation. Biores. Technol. 2006;97:2414–2420. doi: 10.1016/j.biortech.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 15.Geis A.J., Singh R., Teuber M.J. Potential of lactic streptococci to produce bacteriocin. Appl. Environ. Microbiol. 1983;45:205–211. doi: 10.1128/aem.45.1.205-211.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilberg A. Enzyme and bioactive peptides from fish waste related to fish silage, fish feed and fish sauce production. J Aqu. Food Prod. Techonol. 2004;13(2):3–11. [Google Scholar]
- 17.Gonzlez B., Diez V. The effect of nitrite and starter culture on microbiological quality of “chorizo”—a Spanish dry cured sausage. Meat Science. 2002;60:295–298. doi: 10.1016/S0309-1740(01)00137-1. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez C.J., Encinas J.P., Garcia-Lopez M.L., Otero A. Characterization and identification of lacticacid bacteria from fresh water fishes. Food Microbiol. 2000;17:383–391. [Google Scholar]
- 19.Hagi T., Tanaka D., Iwamura Y., Hoshino T. Diversity and seasonal changes in lactic acid bacteria in the intestinal tract of cultured freshwater fish. Aquaculture. 2004;234:335–346. [Google Scholar]
- 20.Halami P.M., Ramesh A., Chandrashekhar A. Fermenting cucumber, a potential source for the isolation of pediocin-type Bacteriocin producer. World J. Microbiol. Biotechnol. 2005;21:1351–1358. [Google Scholar]
- 21.Healy M., Green A., Healy A. Bioprocessing of Marine Crustacean Shell Waste. Acta Biotechnol. 2003;23:151–160. [Google Scholar]
- 22.Holt J.G., Krieg N.R., Sneath P.H.A., Staley J.T., Williams S.T. Bergey’s manual of determinative bacteriology. 9th. William and Wikkins, MD; 1994. pp. 559–564. [Google Scholar]
- 23.Kalalou I., Faid M., Ahami A.T. Extending shelf life of fresh minced camel meat at ambient temperature by Lactobacillus dlbrueckii subsp. Delbrueckii. 2004;7:1–6. [Google Scholar]
- 24.Ndaw A.D., Faid M., Bouseta A., Zinedine A. Effect of controlled lactic acid bacteria fermentation on the microbiological and chemical quality of Moroccan sardines (Sardine pilchardus) Int. J. Agri. Biol. 2008;10:21–27. doi: 10.1556/AMicr.55.2008.3.2. [DOI] [PubMed] [Google Scholar]
- 25.Nair P.S., Surendran P.K. Biochemical characterization of lactic acid bacteria isolated from fish prawn. J. Culture collection. 2005;4:48–52. [Google Scholar]
- 26.Rao M.S., Stevens W.F. Fermentation of shrimp biowaste under different salt concentrations with amylolytic and non-amylolytic lactobacillus strains for chitin production. Food Technol. Biotechnol. 2006;4:83–87. [Google Scholar]
- 27.Ringo E., Bendiksen H.R., Wesmajervi M.S., Olsen R.E., Jansen P.A., Mikkelsen H. Lactic acid bacteria associated with digestive tract of Atlantic salmon (Salmo salar L.) J. Appl. Microbiol. 2000;89:317–322. doi: 10.1046/j.1365-2672.2000.01116.x. [DOI] [PubMed] [Google Scholar]
- 28.Rustad T. Utilization of marine by-products. Electronic J. Environ. Agric. Food Chem. 2003;2:458–463. [Google Scholar]
- 29.Sambrook J., Russell D.W. 3rd. New York, USA: Cold Spring Harbor Laboratory Press; 2001. Molecular Cloning- A laboratory manual. [Google Scholar]
- 30.Sachindra N.M., Bhaskar N. In-vitro antioxidant activities of liquor from fermented shrimp biowaste. Biores. Technol. 2008;99:9013–9016. doi: 10.1016/j.biortech.2008.04.036. [DOI] [PubMed] [Google Scholar]
- 31.Shirai K., Guerrero I., Huerta S., Saucedo G., Castillo A., Gonzalez R. O., Hall G.M. Effect of initial glucose concentration and inoculation level of lactic acid bacteria in shrimp waste ensilation. Enz. Micro. Technol. 2001;28:446–452. doi: 10.1016/s0141-0229(00)00338-0. [DOI] [PubMed] [Google Scholar]
- 32.Sudeepa E.S., Rashmi H.N., Tamil Selvi A., Bhaskar N. Proteolytic bacteria associated with fish processing waste: Isolation and Characterization. J. Food Sci. Technol. 2007;44(3):281–284. [Google Scholar]
- 33.Sudeepa E.S., Tamil Selvi A., Bhaskar N. Antimicrobial activity of lipolytic and hydrocarbon degrading lactic acid bacterium isolated from fish processing waste. J. Food Sci. Technol. 2007;44(4):417–421. [Google Scholar]
- 34.Tannock G.W. A Special Fondness for Lactobacilli. Appl. Environ. Microbiol. 2004;70:3189–3194. doi: 10.1128/AEM.70.6.3189-3194.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vijayabaskar P., Somasundaram S.T. Isolation of bacteriocin producing lactic acid from fish gut and probitic activity against common fresh water fish pathogen Aeromonas hydrophila. Biotechnology. 2008;7(1):124. [Google Scholar]


