Abstract
For commercial purposes, the winemaking industry is constantly searching for new yeast strains. Historically, this has been achieved by collecting wild strains and selecting the best for industrial use through an enological evaluation. Furthermore, the increasing consumer demands have forced the industry to incorporate new strategies such as genetic engineering to obtain improved strains. In response to the lack of public acceptance of this methodology, alternative strategies based on breeding have gained acceptance in recent years. Through the use of conjugation of individual spores without the support of genetic engineering methods we generated intraspecific hybrids from wild strains with outstanding enological characteristics and interdelta fingerprinting was used to confirm the hybrid condition. A detailed enological characterization of the hybrids in synthetic and natural must indicates that physiological parameters such as sporulation, residual sugar, ethanol yield and total nitrogen uptake are within the levels determined for the parental strains, however, other parameters such as growth rate, lag phase and ethanol production show statistical differences with some parental or commercial strains. These findings allow us to propose these hybrids as new wine-making strains.
Keywords: Yeast, Hybrids, Wine
INTRODUCTION
S. cerevisiae is the main species responsible for alcoholic fermentation due to its capacity to grow in sugar-rich environments and withstand high ethanol concentrations. The physiological characteristics of this yeast directly affect the properties of the wine produced and therefore the selection of yeast strains with outstanding enological characteristics has been an area of great interest (26). The selection strategies have been mainly based on isolating a large number of wild yeasts individually evaluated for each of the phenotypes of interest which entails a long and expensive process. An alternative approach is the use of breeding strategies, which are successfully applied in plant and animal improvement (6, 10). Several authors have shown that various characters of enological importance such as ethanol tolerance, production of volatile acid and H2S are polygenic and inheritable thereby supporting the application of a genetic improvement strategy through breeding (4, 15, 19).
Genetic improvement strategies involving breeding have allowed researchers to obtain strains that flocculate and do not produce H2S from strains of S. cerevisiae that exhibit these phenotypes independently (31). In other studies, intraspecific hybrids with an increased bioethanol production (40) or a greater thermotolerance (21) have been obtained. However, many of the strains produced by intraspecific hybridization have been obtained by incorporating genetic markers through genetic engineering. Therefore, given the experimental difficulties derived from the complex characteristics of the wild yeast sexual cycle and the lack of natural genetic markers directed crosses are favored (14, 20, 24, 29, 31). Likewise, to direct crosses and select hybrids, Marullo et al. (20) genetically manipulated strain SB by mutating the HO gene and generated strains that were crossed in the haploid spore stage. However, these strategies limit the use of natural strains since the markers harboring the desirable enological qualities need to be present and/or because the genetically manipulated strains still have little commercial acceptance.
One of the most important enological factors for the wine industry is the amount of ethanol produced. Strains with a low ethanol yield are required in areas where the grapes have a high sugar content, and thus various strategies have been carried out, for example to increase the amount of glycerol produced (8, 23, 27). Furthermore, kinetic properties such as a short lag phase, high growth rate and the capacity to complete fermentation quickly are also important (2) because these characteristics show the ability of the yeast to adapt to the must and begin the fermentation rapidly.
On the other hand, the use of molecular methods for identification and characterization of yeast strains are widely applied. The most common techniques are pulsed field gel electrophoresis (PFGE) (34), restriction fragment length polymorphism (RFLP) (28) and amplified fragment length polymorphism (AFLP) (38). However, the use of these methods for hybrid identification require the parental strains to have detectable differences at a molecular level making these methods useful in the identification of interspecific hybrids (1, 17, 22).
In this work, we generate intraespecific hybrids from wild strains with outstanding enological properties but without natural markers and/or previous genetic modification. Furthermore, we evaluate the use of molecular methods for the selection of intraspecific hybrids and propose that this experimental strategy will allow the industry to use hybrids directly.
MATERIALS AND METHODS
Yeast strains and culture conditions
The yeasts used in this study were the wild strains L3217, L3218 and their monosporic derivatives. Commercial strains EC1118 (Lallemand) and XL (DSM Food Specialties) were used as controls for fermentation parameters. Monosporic derivatives of strain EC1118 were used for sporulation assays. Yeasts were grown at 28 °C on YPD medium (0.5 % yeast extract, 0.5 % peptone, 2 % glucose) and the media was solidified with 2 % agar when required. The fermentations were carried out in 50 ml of synthetic must (12) at 28 °C or in natural must Chardonnay (Maule, Chile) under white wine fermentation conditions.
Growth curves
The growth curves were carried out in 200 μl microfermentations in synthetic must and the optical density was determined at 630 nm in a multiwell plate reader (Biotek). The analysis of data obtained was done using the Gen5 software (Biotek). Maximum growth rate was determined from the slope of the Biomass v/s Time curve during exponential growth using the Excel software (Microsoft). The lag phase (h) was the time between inoculation and the time when the optical density began to increase. The biomass was determined as described by Salinas et al (33).
Sporulation and hybrid formation
Sporulation was induced on acetate medium (1 % potassium acetate, 0.1 % yeast extract, 0.05 % glucose, 2 % agar) at 28 °C. The sporulation capacity was classified into 3 categories: Fast, when asci were observed within 7 days of culture in sporulation media; Late, when there were no asci after 7 and before 30 days and None when no asci were observed following 30 days of incubation. Crossbreeding was achieved by spore conjugation, as described by Winge and Laustsen (39). Ascus dissection, spore isolation and crossbreeding were performed with a Nikon Eclipse 50i micromanipulator.
Molecular evaluation of the hybrids
The hybrid condition was evaluated by PFGE (34), AFLP (38) and interdelta fingerprinting (16).
Fermentations in natural must
"Pied de Cuve" were made in 400 ml of Chardonnay must previously pasteurized at 60 °C for 10 min and inoculated with 4 x 107 cell/ml of yeast from a culture in YPD. After 24 h, 400 ml were used to inoculate 4 l of the same must containing 30 mg/l sulphurous anhydride supplemented with 90 mg/l of ammonium phosphate. The fermentations were considered finished when the residual sugar was 5 g/l or less.
Enological characterization
The residual sugar and ethanol production at the end of the fermentation were determined by HPLC (Waters Corporation) using a Bio-Rad HPX-87H column (25). Ethanol yield (S/E yield) was the conversion efficiency of sugar into ethanol (30). Total acidity was determined by potentiometry as indicated by Bordeau and Scarpa (3).
Total nitrogen uptake
Total nitrogen uptake was calculated as the sum of nitrogen uptake from the amino acids and the nitrogen uptake from ammonium phosphate. To determine the concentration of ammonium phosphate in the fermented must, a rapid enzyme assay from Amonio Megazyme® was used. The sample analysis was performed through a multiwell plate reader (Biotek) using the Gen5 software (Biotek). The nitrogen uptake from ammonium phosphate was defined as the difference between the initial and final amount following fermentation. The concentration of amino acids in the fermented must was determined by HPLC, according to the method described by Janssen et al. (11). The samples from natural must were previously hydrolyzed with HCl 6 N and 1 % phenol for 1 h at 150 °C. The nitrogen uptake from the amino acids was calculated by multiplying the amount of each amino acid consumed by the percentage of nitrogen content in each amino acid.
Statistical analysis
Data were subjected to an analysis of variance (ANOVA) and the mean values of the experiments were statistically analyzed using the LSD test. Differences were considered significant when the probability was < 0.05.
RESULTS
Selection and characterization of parental strains
The wild strains L3217 and L3218 were collected in the Valle del Maipo, Chile, and were selected for their outstanding enological properties for wine production. Since breeding processes require meiotic cells, both strains were sporulated and the cultures from the individual meiotic spores were evaluated in synthetic must. These cultures were subsequently characterized for maximum growth rate, lag phase, residual sugar, S/E yield, nitrogen uptake and ethanol production (Table 1). A similar analysis was done for strains EC1118 and XL, strains normally used in the winemaking industry.
Table 1.
Enological properties of commercial, native, derived monosporic and hybrids strains in synthetic must1.
| Yeast Strain | Total nitrogen uptake (mg/l)2 | Residual sugar (g/l)3 | Ethanol production (%vol) | S/E yield4 | Lag phase (h) | μmax (h-1)5 |
|---|---|---|---|---|---|---|
| L3217 | 121.4 ± 12.6ab | 5.42 ± 0.1bc | 10.4 ± 0.6a | 16.9 ± 1.5d | 11.9 ± 1.6de | 0.17 ± 0.05ab |
| A1 L3217 | 145.2 ± 24.7def | 5.40 ± 0.1bc | 10.7 ± 0.6a | 16.8 ± 0.6d | 12.8 ± 1.6ef | 0.17 ± 0.02a |
| A2 L3217 | 149.0 ± 16.2ef | 5.00 ± 0.3b | 10.3 ± 1.1bcde | 17.1± 1.7abc | 10.1 ± 1.6bcd | 0.16 ± 0.03ab |
| A3 L3217 | 132.9 ± 7.0bcde | 5.60 ± 0.6bc | 10.7 ± 0.3abc | 16.6 ± 0.1cd | 12.8 ± 1.6ef | 0.14 ± 0.04a |
| A11 L3217 | 120.8 ± 11.0ab | 5.45 ± 0.2bc | 10.3 ± 0.4ab | 16.7 ± 0.6cd | 10.1 ± 1.6bcd | 0.15 ± 0.04a |
| A12 L3217 | 127.8 ± 18.9abcd | 5.35 ± 0.2bc | 11.1 ± 0.1abcde | 15.9 ± 0.2abcd | 11.9 ± 1.6de | 0.18 ± 0.04abcd |
| A13 L3217 | 156.7 ± 9.4f | 5.30 ± 0.1bc | 11.3 ± 0.3de | 15.4 ± 0.6ab | 8.5 ± 0.1b | 0.19 ± 0.05abcd |
| L3218 | 109.2 ± 21.7a | 4.95 ± 0.3ab | 11.4 ± 0.2cde | 15.6 ± 0.1abc | 10.1 ± 1.6bcd | 0.17 ± 0.04abc |
| A1 L3218 | 121.2 ± 9.7abc | 4.90 ± 0.1ab | 11.2 ± 0.5bcde | 15.7 ± 1.0abc | 9.2 ± 1.6bc | 0.28 ± 0.06e |
| A2 L3218 | 140.2 ± 8.4bcdef | 4.73 ± 0.1ab | 11.7 ± 0.3de | 15.2 ± 0.1ab | 8.25 ± 0.1b | 0.22 ± 0.04cde |
| A3 L3218 | 149.9 ± 18.8def | 3.80 ± 1.9a | 11.0 ± 0.5cde | 15.7 ± 0.5abc | 11 ± 0.1cde | 0.24 ± 0.04de |
| L3037 | 134.6 ± 9.8bcde | 4.80 ± 0.1ab | 10.7 ± 0.3abcd | 16.3 ± 0.3abcd | 5.5 ± 0.1a | 0.35 ± 0.09f |
| L3038 | 125.3 ± 14.6abc | 4.96 ± 0.1ab | 11.1 ± 0.7abcde | 15.7 ± 0.6abcd | 11.0 ± 0.1cde | 0.22 ± 0.05bcd |
| L3039 | 132.4 ± 10.4bcde | 4.90 ± 0.1ab | 11.0 ± 0.2abcde | 15.9 ± 0.2abcd | 14.7 ± 1.6f | 0.21 ± 0.04bcd |
| EC1118 | 138.9 ± 19.4bcdef | 6.25 ± 0.3c | 11.4 ± 0.2de | 15.2 ± 0.1ab | 10.1 ± 1.6bcd | 0.19 ± 0.03abcd |
| XL | 139.6 ± 17.8cdef | 5.35 ± 0.1bc | 11.5 ± 0.1e | 15.1 ± 0.1a | 11 ± 0.1cde | 0.18 ± 0.03abcd |
Mean of duplicate. Numbers with different letters within a same column differ at p≤0.05 level.
The total nitrogen uptake corresponds to the sum of the nitrogen from the amino acids and ammonium phosphate.
The residual sugar data correspond to the mean differences between the initial and residual concentration of the fermentable sugars.
The S/E yield was calculated based on the amounts of residual sugar and ethanol production under these conditions.
The maximum velocity was obtained from the slope of the exponential phase of the growth curves (Biomass vs Time).
Ideally, yeast used in the wine making industry should have low nitrogen demands. In this sense, the monosporic cultures A11L3217, A12L3217 and A1L3218 showed the lowest total nitrogen uptake in completed fermentations (Table 1). Furthermore, a low efficiency in converting sugar to ethanol is also a desirable characteristic in wine yeasts, particularly in areas where grapes have high sugar levels, since they could produce wines with adequate alcohol levels. This parameter was evaluated at 20 days of fermentation in musts with less than 5 g/l of residual sugars and showed that strains A2L3217 and A11L3217 were the lowest ethanol producing strains.
On the other hand, the kinetic parameters evaluated show that strains A1L3218 and A2L3218 have short lag phases and high growth rates, characteristics that would allow them to quickly dominate the fermentation and impose their physiological properties in the culture. Finally, we evaluated the capacity of the wild strains and their cultures derived from meiotic spores to sporulate. Results shown in Table 2 indicate that all monosporic cultures have lower spore viability than strains L3217 and L3218 with the exception of the culture derived from spore A3L3218 which does not sporulate and spores from strain A2L3218 which have a greater viability than the parental strain. The monosporic cultures derived from strain EC1118 did not sporulate (Table 2), suggesting that these spores are haploid and strain EC1118 is heterozygous for the gene HO (13). The monosporic derivatives that showed high percentages of viability were A2L3217 and A11L3217.
Table 2.
Sporulation time and viability of spore of hybrids and parentals.
| Strain | Sporulation time (d)1 | N0 ascus | Viable spore (%) |
|---|---|---|---|
| L3217 | Fast | 6 | 100 |
| A1L3217 | Fast | 6 | 72.2 |
| A2L3217 | Fast | 6 | 75.0 |
| A3L3217 | Fast | 6 | 72.2 |
| A11L3217 | Fast | 6 | 88.9 |
| A12L3217 | Fast | 6 | 63.2 |
| A13L3217 | Fast | 6 | 61.1 |
| L3218 | Late | 6 | 66.7 |
| A1L3218 | Late | 6 | 66.7 |
| A2L3218 | Late | 6 | 77.8 |
| A3L3218 | None | 0 | ND2 |
| EC1118 | Fast | 6 | 72.2 |
| 1C EC1118 | None | 0 | ND2 |
| 1D EC1118 | None | 0 | ND2 |
| 1E EC1118 | None | 0 | ND2 |
| XL | Fast | 6 | 88.9 |
| L3037 | Late | 6 | 61.1 |
| L3038 | Late | 6 | 55.6 |
| L3039 | Late | 6 | 66.7 |
Sporulation (days): fast (0-7), late (10-30), none (>30).
ND, no determined.
Thus, from the analysis of the enological and biological characteristics of the cultures derived from meiotic spores, we selected strains A2L3218 and A11L3217 as parentals for the hybrid construction.
Obtaining Hybrids
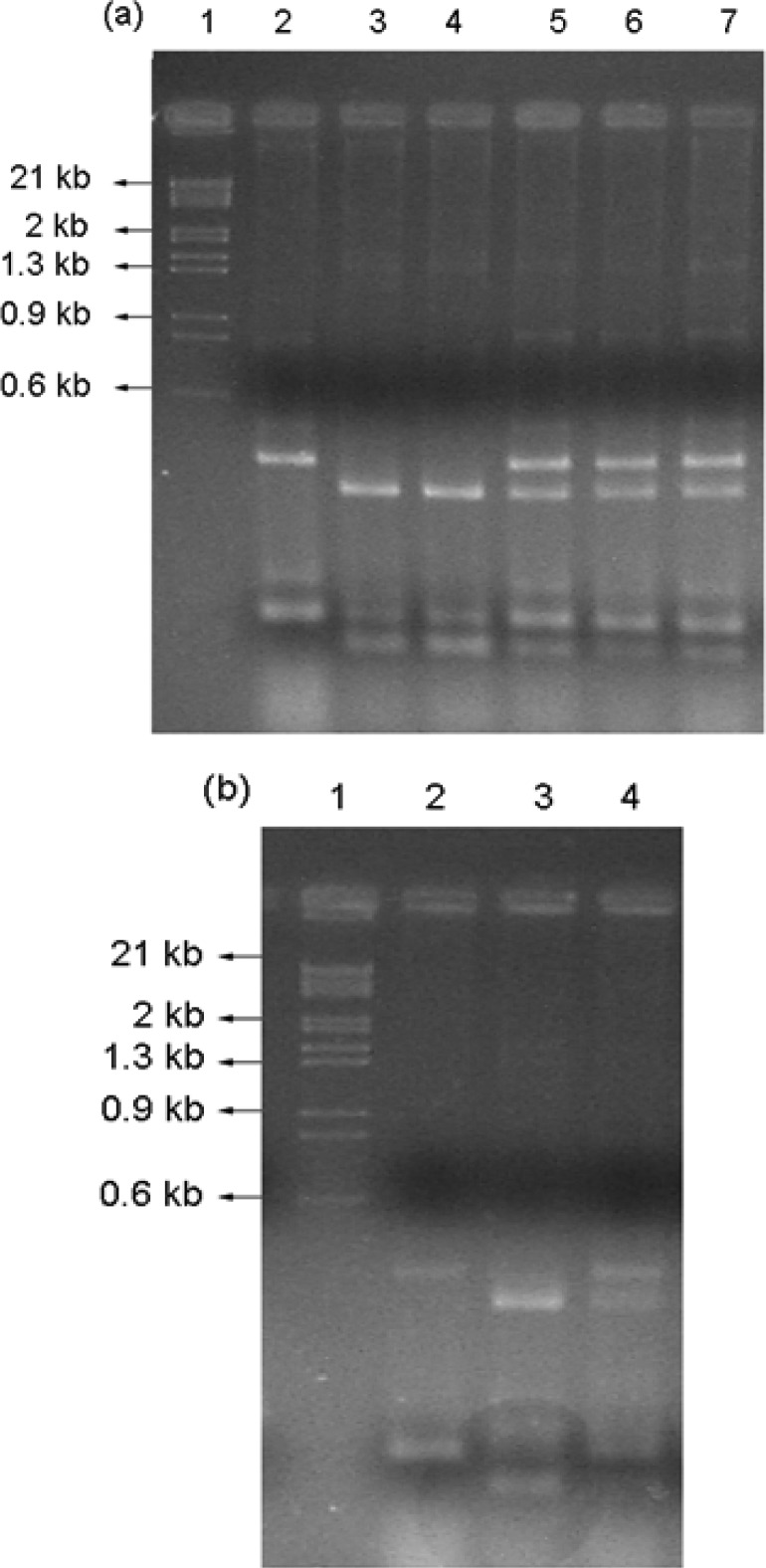
Three hybrids were obtained from the breeding of strains A11L3217 and A2L3218 denominated L3037, L3038 and L3039. To confirm that these strains were hybrids, we performed AFLP, PFGE and interdelta fingerprinting studies between parental strains. The analyses of the electrophoretic patterns from AFLP and PFGE did not differentiate the hybrids from their parentals (data not shown). However, the interdelta fingerprinting analyses detected differences between parental strains and identified the hybrid strain through the presence of bands in each of the parental strains also present in the hybrid strains (Figure 1).
Figure 1.
Verification of hybrid strains by interdelta fingerprinting. (a): Verification of hybrid L3037, L3038 and L3039. Lane 1: Molecular weight standard phage λ DNA digested with EcoRI/HindIII. Lane 2: Parental strain A2L3218 profile, lane 3: A2L3217, lane 4: A11L3217, lane 5-7: Hybrid profiles L3037, L3038 and L3039. (b): Verification of hybrid L3044. Lane 1: Molecular weight standard phage λ DNA digested with EcoRI/HindIII, lane 2: parental A2L3218, lane 3: A2L3217, lane 4: hybrid L3044.
On the other hand, when the capacity of these hybrids to sporulate was analyzed it showed that they all have a late sporulation with almost 60 % viability of spores (Table 2).
Hybrid characterization in synthetic must
Fermentations in synthetic must were carried out by the hybrid strains L3037, L3038 and L3039 and the results are summarized in Table 1. As expected, the values for some parameters such as residual sugar, S/E yield, total nitrogen uptake and ethanol production in the hybrids were found to be between those observed for parental strains. The sugar analyses showed there was less than 5 g/l of sugar remaining, indicating that the hybrids and parental strain A2L3218 were all capable of completing the fermentation. Furthermore, in terms of the nitrogen uptake, the data indicate that all the hybrids analyzed had a similar behavior to the parental strain A11L3217 that is characterized by a preference for amino acids (data not shown).
On the other hand, the kinetic parameters show that the three hybrids had a higher growth rate than the parental strains and of these, hybrid L3037 had the best performance with the highest growth rate and the shortest lag phase of all strains evaluated (Table 1).
Hybrid characterization in natural Chardonnay must
The analyses of the wines obtained from Chardonnay musts fermented with parental and hybrid strains confirm the results obtained in synthetic must, showing that the hybrids L3037 and L3039 produced less ethanol than the commercial strain EC1118 (Table 3).
Table 3.
Enological properties of commercial, parentals and hybrids strains in Chardonnay must.1
| Yeast strain | Total nitrogen uptake2 (mg/l) | Ethanol production (% vol) | Total acidity (g/l of acetic acid) |
|---|---|---|---|
| A2L3218 | 44.7 ± 2.5a | 12.0 ± 0.9 ab | 5.8 ± 0.1b |
| A11L3217 | 78.8 ± 1.2bc | 13.9 ± 2.6 ab | 6.0 ± 0.5b |
| L3037 | 74.5 ± 1.3b | 11.4 ± 0.2 a | 4.6 ± 0.2ª |
| L3038 | 83.6 ± 2.3c | 12.1 ± 0.4 ab | 4.9 ± 0.1ª |
| L3039 | 76.9 ± 1.7bc | 11.3 ± 0.2 a | 4.6 ± 0.1ª |
| EC1118 | 107.6 ± 3.8d | 14.9 ± 2.2 b | 5.0 ± 0.4a |
Numbers with different letters within a same column differ at p≤0.05 level
The total nitrogen uptake corresponds to the sum of the nitrogen from the amino acids and ammonium phosphate.
DISCUSSION
Many authors have obtained hybrids for wine yeasts using micromanipulation as a breeding strategy (5, 14, 20, 21, 24, 31, 39), however, in the majority of these works genetic markers have been used to overcome the difficulties presented by the experimental handling of homothallism in wild yeasts and thus identify the hybrids formed (14, 20, 24, 29). In this work, we generated intraspecific hybrids from natural isolates of S. cerevisiae with interesting enological properties without the use of genetic markers.
Initially, we evaluated several molecular characterization techniques such as PFGE and AFLP widely used in yeast. The electrophoretic profiles detected by these techniques did not allow a clear differentiation between the strains used possibly because the strains that are collected in the same geographic region show a similar genomic constitution (9, 18, 37). However, interdelta fingerprinting was an adequate technique to identify differences between parental strains as well as between the hybrids obtained and their parental strains. Interdelta fingerprinting is effective for the analysis and comparison of the yeast genome, since it consists of amplifying "interdelta" regions or Long Terminal Repeats (LTR) that flank retrotransposons Ty1 and Ty2 present in several copies in yeasts (16) and is a methodology that has been successful in inter and intraspecific discrimination. This technique was used to differentiate S. cerevisiae, Zygosaccharomyces bailii and Saccharomycodes ludwigii responsible for the refermentation of wine during aging in spite of showing a tight relationship as described by their karyotypic analysis (7). Furthermore, it was also able to unequivocally differentiate between 53 commercial yeast strains used in laboratories and wild isolates of S. cerevisiae (16).
On the other hand, the characterization of the fermentations carried out by the parental and hybrid strains indicated that some of the parameters evaluated in the hybrids, such as ethanol production, residual sugar and S/E yield showed values between both parentals. This indicates that the phenotypes in the hybrid strains were affected by the alleles of both parentals, generating intermediate phenotypes as reported by others (20, 35). Interestingly, the kinetic parameters evaluated in synthetic must in the hybrid L3037 show that it has a greater growth velocity and a shorter lag phase than the parental and industrial strains, suggesting a short adaptation time by the yeast to the must. Furthermore, evaluation of this hybrid in natural must indicates that it has a low ethanol production and less nitrogen requirements than the industrial strain suggesting that this strain requires less nitrogen for an adequate fermentation. This is an important characteristic since the exhaustion of nitrogen sources during fermentation stops yeast growth and therefore the fermentation process (36), and a strain that requires lower levels of nitrogen for growth decreases the probabilities of stuck or sluggish fermentations.
The nitrogen uptake observed in natural must differs from that obtained in synthetic must (Tables 1 and 3). This may be a result of different concentrations of nitrogenated sources available in both musts or differences in the genes expressed by wine yeasts in synthetic and natural must as described by Rossouw and Bauer (32). They found that the differences in gene expression in both media were mainly related to metabolic requirements and media composition.
Our results, along with the high viability of spores observed in the hybrids, confirm that it is possible to generate hybrid yeasts through intraspecific genetic improvement programs without the artificial introduction of genetic markers in the parental strains. Furthermore, the selection of hybrids can be made by interdelta fingerprinting as a method of genetic characterization and the strains obtained would not be subject to the restrictions of use of genetically modified organisms.
Acknowledgments
This research was supported by Fondecyt 1070154 and 1100509. V. Garcia was supported by a DICYT-USACH Postdoctoral Fellowship.
REFERENCES
- 1.Antunovics Z., Irinyi L., Sipiczki M. Combined application of methods to taxonomic identification of Saccharomyces strains in fermenting botrytized grape must. J. Appl. Microbiol. 2005;98:971–979. doi: 10.1111/j.1365-2672.2005.02543.x. [DOI] [PubMed] [Google Scholar]
- 2.Bely M., Sablayrolles J.M., Barre P. Automatic detection of assimilable nitrogen deficiencies during alcoholic fermentation in enological conditions. J Ferment Bioeng. 1990;70:246–252. [Google Scholar]
- 3.Bordeu E., Scarpa J. Análisis químico del vino. Santiago: Pontificia Universidad Católica de Chile; 1998. [Google Scholar]
- 4.Caridi A., Sidari R., Solieri L., Cufari A., Giudici P. Wine colour adsorption phenotype: an inheritable quantitative trait loci of yeasts. J. Appl. Microbiol. 2007;103:735–742. doi: 10.1111/j.1365-2672.2007.03301.x. [DOI] [PubMed] [Google Scholar]
- 5.Coloretti F., Zambonelli C., Tini V. Characterization of flocculent Saccharomyces interspecific hybrids for the production of sparkling wines. Food Microbiol. 2006;23:672–676. doi: 10.1016/j.fm.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 6.Crow J. 90 Years ago: The beginning of hybrid maize. Genetics. 1998;148:923–928. doi: 10.1093/genetics/148.3.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Divol B., Miot-Sertier C., Lonvaud-Funel A. Genetic characterization of strains of Saccharomyces cerevisiae responsible for 'refermentation' in Botrytis-affected wines. J. Appl. Microbiol. 2006;100:516–526. doi: 10.1111/j.1365-2672.2005.02818.x. [DOI] [PubMed] [Google Scholar]
- 8.Eustace R., Thornton R.J. Selective hybridization of wine yeasts for higher yields of glycerol. Can. J. Microbiol. 1987;33:112–117. [Google Scholar]
- 9.Flores E.P., Alba J.F., Arrizon J.P., Romano P., Capece A., Gschaedler Mathis A. The uses of AFLP for detecting DNA polymorphism, genotype identification and genetic diversity between yeasts isolated from Mexican agave-distilled beverages and from grape musts. Lett. Appl. Microbiol. 2005;41:147–152. doi: 10.1111/j.1472-765X.2005.01738.x. [DOI] [PubMed] [Google Scholar]
- 10.Giudici P., Solieri L., Pulvirenti A.M., Cassanelli S. Strategies and perspectives for genetic improvement of wine yeasts. Appl. Microbiol. Biotechnol. 2005;66:622–628. doi: 10.1007/s00253-004-1784-2. [DOI] [PubMed] [Google Scholar]
- 11.Janssen P., van Nispen J., Melgers P., van den Bogaart H., Hamelinck R., Goverde B. HPLC analysis of phenylthiocarbamyl (PTC) amino acids: I. Evaluation and optimization of the procedure. Chromatographia. 1986;22:345–350. [Google Scholar]
- 12.Jiranek V., Langridge P., Henschke P.A. Amino acid and ammonium utilization by Saccharomyces cerevisiae wine yeasts from a chemically defined medium. Am J Enol Vitic. 1995;46:75–83. [Google Scholar]
- 13.Johnston J., Baccari C., Mortimer R.K. Genotypic characterization of strains of commercial wine yeasts by tetrad analysis. Res Microbiol. 2000;151:583–590. doi: 10.1016/s0923-2508(00)00228-x. [DOI] [PubMed] [Google Scholar]
- 14.Jolly N.P., Janse B.J.H., Van Rooyen T.J, Louw J.H. Hybridization and typing of yeasts used in sparkling wine fermentations. Am. J. Enol. Vitic. 1993;44:217–226. [Google Scholar]
- 15.Katou T., Namise M., Kitagaki H., Akao T., Shimoi H. QTL mapping of sake brewing characteristics of yeast. J Biosci Bioeng. 2009;107:383–93. doi: 10.1016/j.jbiosc.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 16.Legras J.L., Karst F. Optimisation of interdelta analysis for Saccharomyces cerevisiae strain characterisation. FEMS Microbiol Lett. 2003;221:249–255. doi: 10.1016/S0378-1097(03)00205-2. [DOI] [PubMed] [Google Scholar]
- 17.Lopandic K., Gangl H., Wallner E., Tscheik G., Leitner G., Querol A., Borth N., Breitenbach M., Prillinger H., Tiefenbrunner W. Genetically different wine yeasts isolated from Austrian vine-growing regions influence wine aroma differently and contain putative hybrids between Saccharomyces cerevisiae and Saccharomyces kudriavzevii. FEMS Yeast Res. 2007;7:953–965. doi: 10.1111/j.1567-1364.2007.00240.x. [DOI] [PubMed] [Google Scholar]
- 18.Martínez C., Gac S., Lavin A., Ganga M. Genomic characterization of Saccharomyces cerevisiae strains isolated from wine-producing areas in South America. J. Appl. Microbiol. 2004;96:1161–1168. doi: 10.1111/j.1365-2672.2004.02255.x. [DOI] [PubMed] [Google Scholar]
- 19.Marullo P., Bely M., Masneuf-Pomarede I., Aigle M., Dubourdieu D. Inheritable nature of enological quantitative traits is demonstrated by meiotic segregation of industrial wine yeast strains. FEMS Yeast Res. 2004;4:711–719. doi: 10.1016/j.femsyr.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 20.Marullo P., Bely M., Masneuf-Pomarede I., Pons M., Aigle M., Dubourdieu D. Breeding strategies for combining fermentative qualities and reducing off-flavor production in a wine yeast model. FEMS Yeast Res. 2006;6:268–279. doi: 10.1111/j.1567-1364.2006.00034.x. [DOI] [PubMed] [Google Scholar]
- 21.Marullo P., Mansour C., Dufour M., Albertin W., Sicard D., Bely M., Dubourdieu D. Genetic improvement of thermo-tolerance in wine Saccharomyces cerevisiae strains by a backcross approach. FEMS Yeast Res. 2009;9:1148–1160. doi: 10.1111/j.1567-1364.2009.00550.x. [DOI] [PubMed] [Google Scholar]
- 22.Massoutier C., Alexandre H.M.F., Charpentier C. Isolation and characterization of cryotolerant Saccharomyces strains. Vitis. 1998;37:55–59. [Google Scholar]
- 23.Michnick S., Roustan J.L., Remize F., Barre P., Dequin S. Modulation of glycerol and ethanol yields during alcoholic fermentation in Saccharomyces cerevisiae strains overexpressed or disrupted for GPD1 encoding glycerol 3-phosphate dehydrogenase. Yeast. 1997;13:783–793. doi: 10.1002/(SICI)1097-0061(199707)13:9<783::AID-YEA128>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 24.Nakazawa N., Iwano K. Efficient selection of hybrids by protoplast fusion using drug resistance markers and reporter genes in Saccharomyces cerevisiae. J Biosci Bioeng. 2004;98:353–358. doi: 10.1016/S1389-1723(04)00295-6. [DOI] [PubMed] [Google Scholar]
- 25.Nissen T.L., Schulze U., Nielsen J., Villadsen J. Flux distributions in anaerobic, glucose-limited continuous cultures of Saccharomyces cerevisiae. Microbiology. 1997;143:203–218. doi: 10.1099/00221287-143-1-203. [DOI] [PubMed] [Google Scholar]
- 26.Pretorius I.S. Tailoring wine yeast for the new millennium: novel approaches to the ancient art of winemaking. Yeast. 2000;16:675–729. doi: 10.1002/1097-0061(20000615)16:8<675::AID-YEA585>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 27.Prior B.A., Baccari C., Mortimer R.K. Selective breeding of Saccharomyces cerevisiae to increase glycerol levels in wine. J. Int. Sci. Vign. Vin. 1999;33:57–65. [Google Scholar]
- 28.Querol A., Barrio E., Huerta T., Ramon D. Molecular monitoring of wine fermentations conducted by active dry yeast strains. Appl. Environ. Microbiol. 1992;58:2948–2953. doi: 10.1128/aem.58.9.2948-2953.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramirez M., Perez F., Regodon J.A. A simple and reliable method for hybridization of homothallic wine strains of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 1998;64:5039–5041. doi: 10.1128/aem.64.12.5039-5041.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ribereau-Gayon P., Dubourdieu D., Doneche B., Lonvaud A. Handbook of enology. New York: Jonh Wiley and Sons; 2000. [Google Scholar]
- 31.Romano P., Soli M.G., Suzzi G., Grazia L., Zambonelli C. Improvement of a wine Saccharomyces cerevisiae strain by a breeding program. Appl. Environ. Microbiol. 1985;50:1064–1067. doi: 10.1128/aem.50.4.1064-1067.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rossouw D., Bauer F.F. Comparing the transcriptomes of wine yeast strains: toward understanding the interaction between environment and transcriptome during fermentation. Appl. Microbiol. Biotechnol. 2009;84:937–954. doi: 10.1007/s00253-009-2204-4. [DOI] [PubMed] [Google Scholar]
- 33.Salinas F., Mandakovic D., Urzua U., Massera A., Miras S., Combina M., Angelica Ganga M., Martínez C. Genomic and phenotypic comparison between similar wine yeast strains of Saccharomyces cerevisiae from different geographic origins. J Appl Microbiol. 2010;108:1850–1858. doi: 10.1111/j.1365-2672.2010.04689.x. [DOI] [PubMed] [Google Scholar]
- 34.Schwartz D.C., Cantor C.R. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell. 1984;37:67–75. doi: 10.1016/0092-8674(84)90301-5. [DOI] [PubMed] [Google Scholar]
- 35.Shinohara T., Saito K., Yanagida F., Goto S. Selection and hybridization of wine yeasts for improved winemaking properties: Fermentation rate and aroma productivity. J. Ferment. Bioeng. 1994;77:428–431. [Google Scholar]
- 36.Varela C., Pizarro F., Agosin E. Biomass content governs fermentation rate in nitrogen-deficient wine musts. Appl Environ Microbiol. 2004;70:3392–3400. doi: 10.1128/AEM.70.6.3392-3400.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vezinhet F., Hallet J.-N., Valade M., Poulard A. Ecological survey of wine yeast strains by molecular methods of identification. Am. J. Enol. Vitic. 1992;43:83–86. [Google Scholar]
- 38.Vos P., Hogers R., Bleeker M., Reijans M., van de Lee T., Hornes M., Frijters A., Pot J., Peleman J., Kuiper M. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winge O., Laustsen O. Artificial species hybridization in yeast. C R Trav Lab Carlsberg Ser Physiol. 1938;22:235–244. [Google Scholar]
- 40.Zheng D.Q., Wu X.C., Tao X.L., Wang P.M., Li P., Chi X.Q., Li Y.D., Yan Q.F., Zhao Y.H. Screening and construction of Saccharomyces cerevisiae strains with improved multi-tolerance and bioethanol fermentation performance. Bioresour. Technol. 2011;102:3020–3027. doi: 10.1016/j.biortech.2010.09.122. [DOI] [PubMed] [Google Scholar]