Abstract
Shewanella are renowned for their ability to respire on a wide range of electron acceptors, which has been partially accredited to the presence of a large number of the c-type cytochromes. In the model species S. oneidensis MR-1, at least 41 genes encode c-type cytochromes that are predicted to be intact, thereby likely functional. Previously, in-frame deletion mutants for 36 of these genes were obtained and characterized. In this study, first we completed the construction of an entire set of c-type cytochrome mutants utilizing a newly developed att-based mutagenesis approach, which is more effective and efficient than the approach used previously by circumventing the conventional cloning. Second, we investigated the cytochrome c maturation (Ccm) system in S. oneidensis. There are two loci predicted to encode components of the Ccm system, SO0259-SO0269 and SO0476-SO0478. The former is proven essential for cytochrome c maturation whereas the latter is dispensable. Unlike the single operon organization observed in other γ-proteobacteria, genes at the SO0259-SO0269 locus are uniquely organized into four operons, ccmABCDE, scyA, SO0265, and ccmFGH-SO0269. Functional analysis revealed that the SO0265 gene rather than the scyA and SO0269 genes are relevant to cytochrome c maturation.
Introduction
Cytochromes, proteins carrying the heme group as prosthetic cofactor, can be classified into a-, b-, c-, d-, and o-types depending on the variations on the protoporphyrin ring [1]. Cytochromes of c-type, existing as membrane-bound proteins or soluble periplasmic proteins, play vital roles in bacterial respiration and photosynthesis as enzymes to exchange electrons with the bound substrates or as pure electron carriers to shuttle electrons. It is estimated that one third of cytochrome hemes are located at enzyme active sites while the rest are redox cofactors transporting electrons in an electron transfer chain [1–3]. The unique feature of c-type cytochromes is the covalent attachment of the cofactor to the protein polypeptide at the cysteines within the signature haem c binding motif (HBM) CX nCH (X stands for any amino acid, n = 3, 4, 15) [1–3]. Although the motif is well conserved across species, the sequences A/FX 2CH have been described as HBMs in bacterial or mitochondrial proteins [4,5].
Shewanella oneidensis MR-1, a facultative Gram-negative anaerobe, is renowned for its remarkable anaerobic respiration ability. Linked to this unique characteristic is a high cytochrome content, especially c-type [6,7]. Compared to Escherichia coli which hosts only 5~7 c-type cytochromes, S. oneidensis is predicted to possess as many as 44 c-type cytochrome proteins by screening for the canonical HBMs in the proteome [8–10]. Although a few appear to be degenerated due to frameshift mutations within the coding sequence [9], the number of c-type cytochromes in S. oneidensis may increase with time as proteins with the non-canonical HBMs may be found. For example, haem c group II in OTR (SO4144, octaheme tetrathionate reductase) is ligated to C74X75X76C77H78 and a lysine residue (Lys56), which are in proximity structurally [11,12].
Extensive biochemical and genetic investigations have revealed that three systems predominate in c-type cytochrome maturation although some specialized ones have been identified in recent years [1,13,14]. The definition of the three systems is based on the presence of specific assembly components that are unique to each maturation system. System I, also called Ccm (cytochrome c maturation), extensively studied in α and γ-proteobacterial models, is composed of up to 12 components, CcmA to CcmH, CcmI, DsbA, DsbB, and DsbD/CcdA. As a γ-proteobacterium, S. oneidensis is predicted to have system I as it encodes analogues to CcmC, CcmF, and CcmE, the signature components for this system [1,6]. However, this organism differs from other γ-proteobacteria in ccm gene organization significantly [15]. Unlike the common pattern that all ccm genes are clustered together and transcribed in the same orientation, in S. oneidensis two genes separate ccmABCDE from ccmFGH, resulting in two ccm operons, which are transcribed divergently. In addition, it is interesting to note that this microorganism has a second ccmF gene located elsewhere on the chromosome.
In S. oneidensis, the functionally defined c-type cytochromes mostly are terminal reductases as their corresponding deletion mutants display distinguishable phenotypes, such as NrfA, NapB (small subunit of nitrate reductase), FccA (fumarate reductase), and DmsE (subunit of dimethyl sulfoxide (DMSO) reductase), to name a few [13–15]. Furthermore, those involved in respiration of insoluble electron acceptors are relatively better understood because the subject has been under intensive investigation for nearly three decades [7,16]. To facilitate systematic characterization of c-type cytochromes, we endeavored to construct a whole set of single-gene knockouts but failed with five of them in our previous study [10]. Here, we first developed an att-based mutagenesis approach and completed the construction of an entire set of c-type cytochrome mutants with it. This new approach bypasses the conventional cloning step, which reduces the effectiveness and efficiency of the system used before. In addition, we examined the Ccm system for determination of the essential components of the system. More intriguing, this research identifies a protein encoded in the ccm locus showing varying degrees of essentialness for respiration of different electron acceptors.
Results
Activeness of c-type cytochrome genes at transcriptional levels
Recently, proteomic measurements reveal 23 of all S. oneidensis c-type cytochromes in cells grown with ferric citrate (soluble Fe(III)) or MnO2 (insoluble Mn(IV)) as electron acceptors under anaerobic conditions. This observation implicates that production of a considerable number of c-type cytochromes in physiologically relevant amounts is condition-specific [17]. Such information not only provides insights into their physiological functions but also may help to understand technological difficulties encountered during construction of a whole set of c-type cytochrome single-gene mutants. In our previous study, we failed to generate in-frame deletions for scyA (SO0264), torC (SO1233), SO1748, ccpA (SO2178) or SO3056 after multiple attempts [10].
To understand expression characteristics of c-type cytochromes in cells grown under aerobic and anaerobic conditions, we examined the mRNA abundance of their coding genes in exponentially growing cells using quantitative reverse transcription PCR (qRT-PCR) (Figure 1). The cymA gene, as reported repeatedly [18,19], showed a constant high level of transcription that is oxygen-independent. This is not surprising because the protein plays a key role in mediating electron transport in multiple respiratory pathways [20,21]. Interestingly, impacts of oxygen on transcription of the major components of the metal reduction pathway mtrA, mtrC, and omcA were also negligible. Combining growth defects of the ∆mtrA and ∆mtrC strains under aerobic conditions [10], these data suggest that these proteins and/or the pathway may be implicated in other physiological processes of general importance. Consistent with findings that the cbb 3 oxidase plays a predominant role in oxygen respiration [22], both ccoP and ccoO (encoding two essential subunits of the cbb 3 oxidase) were transcribed at significantly higher levels in cultures grown with oxygen than in those without oxygen. Other genes displaying the same pattern include scyA, cctA, and cytcB, implicating that these small electron shuttling c-type cytochromes may be more likely to function under aerobic conditions. In contrast, fccA (fumarate reductase) and dmsE (subunit of the DMSO reductase) were transcribed at higher levels in anaerobic than aerobic cultures. It is worth mentioning that transcription of a number of genes, most of which encode proteins of unknown function, was extremely low regardless of growth conditions. While one of possible explanations is that the culturing conditions do not favor transcription of these genes, at least one alternative has been reported. In S. oneidensis, the caa 3 oxidase has a negligible role in oxygen respiration because its coding operon (including coxB) is not expressed to a physiologically relevant level [22].
Figure 1. Expression of genes encoding c-type cytochromes in S. oneidensis.
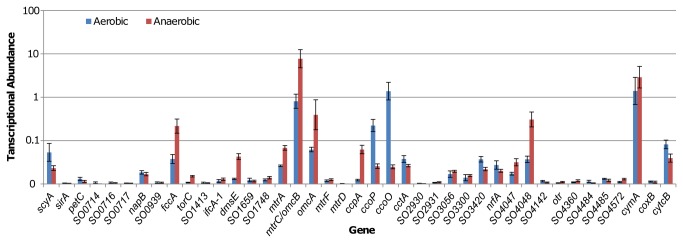
qRT-PCR analysis of RNA extracted from mid-logarithmic growing cells (OD600, ~0.4 and ~0.15 for aerobic and anaerobic cultures respectively). All data were normalized to expression of the arcA and recA genes, which were constant during the exponential growth phase. Numbers reported are standardized to expression of the arcA gene, but the same trends are observed when standardized to expression of the recA gene. Error bars represent standard deviation for triplicate cultures. SO1778 is displayed as mtrC/omcB because both names have been used in previous publications, but only mtrC is used in the rest of this article since it is accepted more widely.
Development of an att-based mutagenesis system and generation of a complete set of S. oneidensis c-type cytochrome mutants
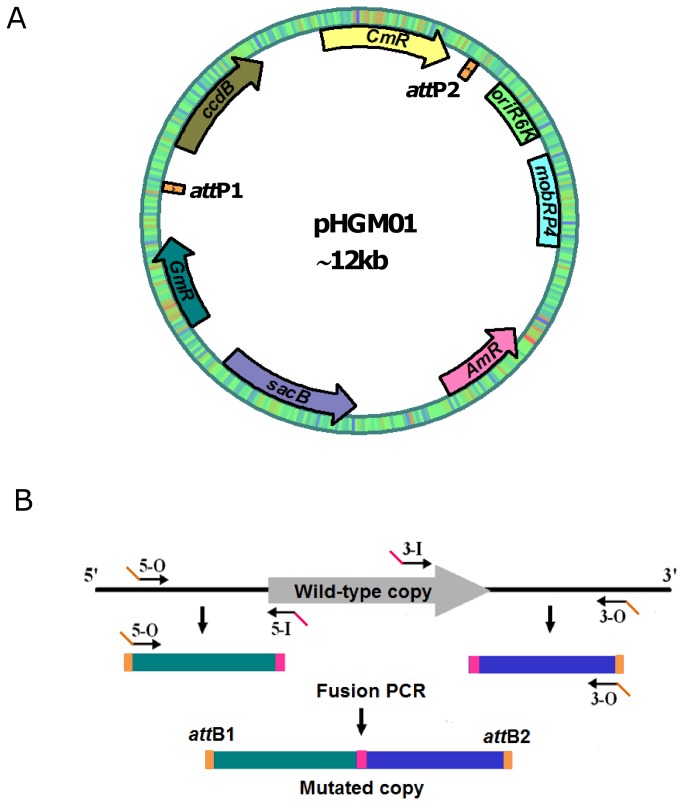
In S. oneidensis, in-frame single-gene deletion strains were successfully constructed for 36 out of 41 c-type cytochrome genes using either the fusion-PCR- or cre-lox-based mutagenesis approach [10]. Among five c-type cytochrome genes that we failed to remove, only scyA was highly expressed aerobically as shown above, a condition under which the previous attempts are made [10], suggesting that most of these genes may not play an important role in aerobiosis. In addition, these genes do not carry common characteristics in sequences and possible secondary structures that may interfere with PCR amplification or recombination. As a consequence, the failure is likely ascribable to the mutagenesis approaches. Indeed, both the fusion-PCR-based and cre-lox-based methods proved inefficient and sometimes even ineffective because of multiple rounds of PCR and conventional cloning. Additionally, the mutagenesis delivery vector pDS3.0 is rather large (~10 kb), resulting in few unique restriction enzyme sites available for cloning, especially for large open reading frames (ORFs). We therefore sought to develop a new mutagenesis system free of conventional cloning for Shewanella and other organisms in which plasmids with a pR6K origin (ori R6K) could not replicate. A PCR fragment containing bacteriophage lambda attP sequences bracketing the CmR cassette and toxic gene ccdB, generated using pMK2010 as the template, was introduced into pDS3.0, resulting in pHGM01 [23] (Figure 2A). To produce an in-frame deletion construct for ORF of interest, two PCR fragments flanking the ORF were generated using primers containing the attB sequences (outside primer) and linking sequences (inside primers) with the genomic DNA as the template by PCR and then joined together by the second round of PCR with primers containing the attB sequences (fusion PCR step) (Figure 2B). The resulting fusion PCR product was transferred into pHGM01 by lambda BP recombination. It is worth noting that the recombination also occurred effectively in the presence of unwanted smaller DNA fragments, which are common, often inevitable, byproducts at the fusion PCR step and regarded to be the major obstacle for conventional cloning. The correct mutagenesis vector, verified by DNA sequencing, was then transferred into S. oneidensis by conjugation for the subsequent steps of the fusion-PCR-based mutagenesis procedure [24].
Figure 2. The att-based mutagenesis developed in this study.
A. Mutational construct delivery vector pHGM01. The vector was created by introducing into the SacI site of pDS3.0 a fragment containing attP1, ccdB, CmR, and attP2 amplified from pMK2010. B. Generation of a fusion PCR product for BP recombination. Outside primers 5-O and 3-O contain attB1 and attB2 sequences, respectively. Inside primers 5-I and 3-I contain linking sequences respectively, which are complementary to each other.
With this new system, we obtained in-frame deletion strains for the scyA, torC, SO1748, ccpA, and SO3056 genes. Physiological characterization of these five mutants was then carried out to assess impacts of each mutation as previously described [10]. To support growth, oxygen or one of following chemical agents was used as the sole electron acceptor: DMSO, fumarate, trimethylamine N-oxide (TMAO), NaNO3, Fe-Citrate, and MnO2 under anaerobic conditions. Results showed that none of mutations had a statistically significant effect on growth under any test condition except for the ∆torC strain grown on TMAO (Table 1). Consistent with the essentiality of the torECAD operon for TMAO reductase activity [25], the loss of the torC gene prevented S. oneidensis from growing on TMAO.
Table 1. Physiological characterization of mutants constructed in this study a.
| Mutant | Possible function of deleted gene | DMSO | Fumarate | TMAO | NaNO3 | Fe-Citrate | MnO2 | O2 |
|---|---|---|---|---|---|---|---|---|
| HG0264 | Periplasmic monoheme cytochrome c 5 | + | + | + | + | + | + | + |
| HG0265 | Cytochrome c biogenesis protein CcmI | -- | -- | + | -- | -- | -- | + |
| HG0266 | Cytochrome c biogenesis protein CcmF | ─ | ─ | ─ | ─ | -- | -- | -- |
| HG0268 | Cytochrome c biogenesis protein CcmH | ─ | ─ | ─ | ─ | -- | -- | -- |
| HG0269 | Unknown | + | + | + | + | + | + | + |
| HG0478 | Cytochrome c biogenesis protein CcmF2 | + | + | + | + | + | + | -- |
| HG0478-6 | CcmF2-NrfF-SO0476, Unknown | + | + | + | + | + | + | + |
| HG1233 | TMAO reduction TorC | + | + | ─ | + | + | + | + |
| HG1748 | Periplasmic monoheme cytochrome c | + | + | + | + | + | + | + |
| HG2178 | Cytochrome c 5 peroxidase CcpA | + | + | + | + | + | + | + |
| HG3056 | Tetraheme cytochrome c | + | + | + | + | + | + | + |
aGrowth of the wild type and mutant strains was monitored in M1 medium with lactate as electron donor and one of listed chemicals as electron acceptor. Values (growth rate and/or maximum cell density) of mutant strains were normalized to that of the wild type: +, > 75%; --, between 75% and 25%; -: < 25%. Experiments were performed at least three times and standard deviation was less than 10% of values.
In silico analysis of Ccm system of S. oneidensis
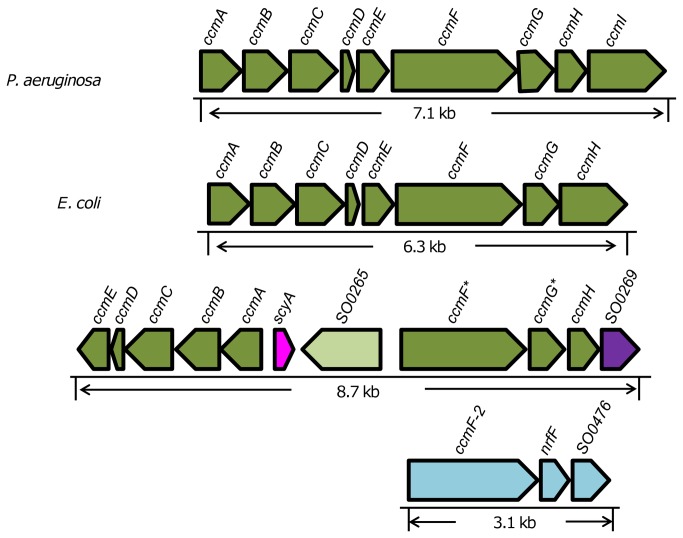
S. oneidensis is distinct from other γ-proteobacteria in organization of the ccm genes (Figure 3). According to the genome annotation, S. oneidensis has two loci for ccm genes, SO0259-0269 and SO0476-0478. The first locus includes operons ccmABCDE, scyA, SO0265, and ccmF 1 -dsbE-ccmH-SO0269. The ccmABCDE operon exists as a single copy in all 27 sequenced Shewanella and the essentiality of their products for cytochrome c maturation have been firmly established [1–3]. Within the ccmF 1 -dsbE-ccmH-SO0269 operon, the dsbE gene (SO0267) has been proposed to encode a real CcmG protein, whose homologues are implicated in the reduction of disulphide bonds of the apocytochrome c prior to haem ligation in a variety of bacteria [15]. This proposal is supported by sequence analysis. SO0267 shares a highest sequence similarity with well-characterized E. coli CcmG proteins: 50%/72% (identity/positive), with an expect value of 2e-38. We therefore renamed dsbE as ccmG, rendering a syntenic consistency to the ccmF 1 GH-SO2069 operon as the order of the ccmFGH(I) genes are perfectly preserved in γ-proteobacteria. However, the last gene of the ccmF 1 GH-SO0269 operon is a mystery. Although the E. coli CcmHEC is 350 amino acids (aa) in length, a size equivalent to a combination of S. oneidensis CcmH (159 aa) and SO0269 (194 aa), SO0269 shows a modest sequence similarity to CcmGEC (32%/52%, 3e-8), but not to CcmHEC. In addition, SO0269 has no sequence similarity to P. aeruginosa CcmIPA (407 aa). Moreover, an analogue of SO0269 is completely missing from two sequenced Shewanella strains, S . denitrificans OS217 and S. violacea DSS12, implying that SO0269 may not be required for cytochrome c maturation. Two remaining genes in this locus are scyA and SO0265. It is interesting to note that SO0265 (415 aa) is a homologue of CcmIPA with sequence similarity of 33%/54% and expect value of 2e-37, implicating a possible role for this protein in cytochrome c maturation.
Figure 3. Organization of the ccm genes in γ-proteobacteria.
The most common gene arrangement is represented by Pseudomonas aeruginosa, in which CcmH and CcmI are separate from each other. The less common one is represented by E. coli, whose CcmH is a fusion protein between CcmH and the C-terminal portion of CcmI found in other bacteria. In S. oneidensis, two loci for predicted ccm genes were shown. Genes renamed in this study were labeled with an asterisk mark. Genes are drawn to scale. The data were from http://img.jgi.doe.gov/cgi-bin/w/main.cgi.
The second locus consists of a single operon, ccmF 2 -nrfF-SO0476, which is perfectly preserved in all sequenced Shewanella . Like CcmF1, CcmF2 is highly homologous to CcmFEC with sequence similarity of 40%/60% and expect value of 2e-137. Such a high level of sequence conservation strongly suggests a close evolutionary relationship between the two proteins. Moreover, NrfF (SO0477) and SO0476 are homologues of CcmH (37%/69%, 2e-16) and SO0269 (38%/63%, 3e-37), respectively.
Maturation of c-type cytochromes is independent of ccmF2-nrfF-SO0476
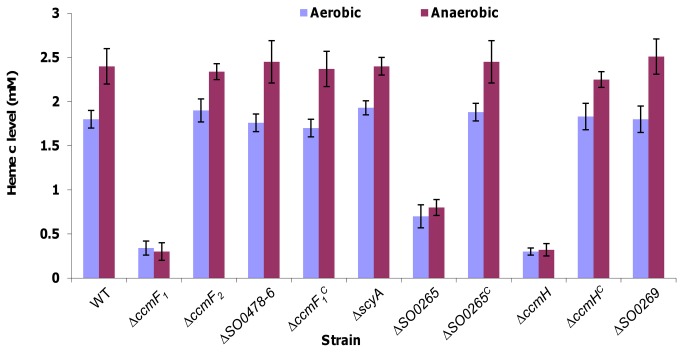
The in silico analysis raised questions about the role played by two CcmF components as well as uncertain SO0265 and SO0269 in S. oneidensis. To evaluate the impact of these proteins on cytochrome c maturation, we constructed mutants defective in one of their coding genes. Deletion of ccmF 2 did not elicit any difference from the parental wild type strain but the loss of ccmF 1 resulted in light-colored colonies, an indicator for the reduced amount of c-type cytochromes. Quantification of intracellular heme c content confirmed that the ∆ccmF 1 strain was severely impaired in heme c production whereas the ∆ccmF 2 strain had a heme level identical to that of the wild type (Figure 4). Importantly, the deficient in heme c resulted from the ccmF1 deletion was corrected by its expression in trans, indicating that the phenotype observed was due to the intended mutation.
Figure 4. Heme c levels in S. oneidensis strains.
wild type, ∆ccmF1, and ∆ccmF2 strains. Mid-logarithmic growing cells (OD600, ~0.4 and ~0.15 under aerobic and anaerobic conditions respectively) were collected for the heme c assay. ∆ccmF 1 c and ∆SO0265 c represents the mutants carrying pHG102-ccmF 1 and pHG102-SO0265, respectively. All other strains carry empty vector pHG102. Three independently collected samples were assayed and the averaged levels were presented in mM per g of proteins. Error bars represent the standard deviations of the data.
To assess impact of loss of the ccmF 1 and ccmF 2 genes on respiration of various electron acceptors, the wild type and mutant strains were inoculated into M1 medium supplemented with one of tested electron acceptors and growth was monitored. With all tested electron acceptors, the wild type and ∆ccmF 2 strains were indistinguishable but the ∆ccmF 1 strain was distinct (Table 1). In the absence of CcmF1, the bacterium lost ability to grow on DMSO, fumarate, TMAO, or NaNO3 completely but was still capable of growing on oxygen, Fe-Citrate, and MnO2, albeit significantly impaired. To examine impact of the entire ccmF 2 -nrfF-SO0476 operon on cytochrome c maturation, we removed all of these three genes from the wild type and characterized the resulting mutants as described above. Similar results were obtained compared to the ∆ccmF 2 strain. These data, collectively, indicate that the ccmF 2 -nrfF-SO0476 operon is not involved in cytochrome c maturation in S. oneidensis. We, therefore, renamed the ccmF 1 gene as ccmF.
Maturation of c-type cytochromes requires SO0265 and CcmH but not SO0269
The scyA gene locates in the middle of the ccm cluster and has been suggested to be involved in cytochrome c maturation [19]. However, our data from the physiological characterization as shown above imply that the protein may not have a role in the process, at least not significantly. This notion is further supported by that the ∆scyA strain had similar levels of heme c production compared to the wild type (Figure 4).
We then made an attempt to determine whether three remaining genes in the cluster, SO0265, ccmH, and SO0269, are relevant to cytochrome c maturation in S. oneidensis. As shown above, the ccmH gene of S. oneidensis is much shorter than its E. coli counterpart. Therefore, whether this protein is essential for cytochrome c maturation merits an investigation. Interestingly, phenotypes resulting from deletions of these genes were different (Figure 4 and Table 1). Apparently, the ccmH gene is totally essential for the process evidenced by that its deletion resulted in a ccmF phenotype. Loss of the SO0265 gene not only caused significant reduction in the bacterial growth on DMSO, fumarate, NaNO3, Fe-Citrate or MnO2, but also impaired growth with oxygen. However, relevance of the gene to TMAO respiration remained to be determined. In line with these growth phenotypes, the SO0265 mutant produced heme c approximately 35% relative to the wild type strain. In contrast, deletion of the SO0269 gene had little impact on aerobic and anaerobic growth as well as heme c production. As a result, whether this protein is part of the Ccm system in S. oneidensis remains to be determined.
Discussion
A new in-frame deletion mutagenesis method, which is based on the site-specific recombination system used by phage λ to integrate its DNA in the E. coli chromosome, has been developed and successfully utilized in this study. To construct Shewanella mutants, fusion PCR technique with various suicide plasmids has been the most frequently utilized [10,24,26,27]. Because of the large size of suicide plasmids and unwanted PCR byproducts, these methods with traditional cut-ligation cloning suffers from the lack of unique restriction sites and/or a low efficiency for ligation of the fusion PCR products. By exploiting the site-specific recombination mechanism, our new system bypasses the need for restriction enzymes and ligation to introduce the fusion PCR products, substantially enhancing cloning efficiency.
The unique organization of the ccm genes is preserved in all sequenced Shewanella strains, implicating a common ancestral linkage. Despite a high level of sequence similarity with CcmF, CcmF2, as well as other proteins encoded by the ccmF 2-nrfF-SO0476 operon, has no role in cytochrome c maturation. Within the genuine ccm cluster, there are two puzzling genes, SO0265 and SO0269. S. oneidensis has an NrfAH type of nitrite reduction system, in which the specific electron transport protein NrfH is replaced by CymA [18]. Given that all NrfA proteins contain an unconventional CXXCK haem-binding motif, which is recognized by a specific haem lyase as in E. coli and Wolinella succinogenes [28–30], it is conceivable that such a lyase exists in S. oneidensis. In a recent report, SO0265 has been suggested to be responsible for haem ligation to the atypical CXXCK site [31]. However, our data indicate that SO0265 certainly plays a more general and important role in cytochrome c maturation as the specific CXXCK lyases are not involved in haem ligation to the conventional sites [28–30]. Given that SO0265 is homologous to CcmIPA, it is likely that SO0265 is a functional equivalence of CcmI, which is currently under study. In the case of SO0269, a question needed to be addressed is whether the protein has a subtle role in cytochrome c maturation or is simply a redundant and degenerated copy of CcmG.
Recently, a few studies have been conducted to elucidate functions of c-type cytochromes that are poorly understood [17,32,33]. We now know that both CctA and FccA, two small abundant periplasmic c-type cytochromes, shuttle electrons from cytoplasmic membrane-bound CymA to outer-membrane-bound MtrA [32]. ScyA, also abundant in the periplasm, is proposed to be the electron donor to the dihaem CcpA, which functions as cytochrome c peroxidase to protect cells from oxidative damage under anaerobic conditions [33]. Together with our physiological analysis, it is apparent that ScyA functions more likely as an electron mediator than as a component of the Ccm system.
Shewanella , along with Geobacter , Anaeromyxobacter and Desulfovibrio , are examples of bacterial genera with a large number of predicted c-type cytochrome genes [34]. It has been suggested that the c-type cytochromome (the total of c-type cytochromes encoded in a given genome) is largely responsible for the respiratory versatility of these microbes. However, most of S. oneidensis c-type cytochromes are missing in the well-characterized model bacteria, such as E. coli, it therefore remains a major challenge to annotate their encoding genes with a specific function. Moreover, the focus of studies on c-type cytochromes of S. oneidensis has been limited to those involved in the anaerobic respiration of various different electron acceptors, especially soluble and insoluble metal oxides [16]. As a result, functions of a large fraction of the entire c-type cytochrome pool remain unknown because many of deletion mutants in c-type cytochrome genes do not have distinct phenotypes under conditions tested [10,35]. This is, to some extent, attributable to the promiscuity of c-type cytochromes given that a large number of proteins sharing similar features coexist in the proteome. For example, terminal reductases OTR and NrfA are able to reduce multiple substrates [12,36]. More importantly, functional redundancy and degeneration of c-type cytochromes may be a common scenario in S. oneidensis. Three of 44 c-type cytochromes predicted initially are either truncated or disrupted, thereby unlikely to be functional [9]. In the case of genes for intact c-type cytochromes, approximately 40% of them are transcribed at extremely low levels and/or their products are not identified in proteomes from cells grown under either aerobic or anaerobic condition, among which the coxB gene (the caa 3 oxidase component) is verified [17,22]. Therefore, to determine physiological roles of unknown c-type cytochromes remains an important task for the future.
Methods
Bacterial strains, plasmids, and growth conditions
A list of all bacterial strains and plasmids used in this study is given in Table 2. Information of primers used for PCR amplification in this study is available upon request. For genetic manipulations, E. coli and S. oneidensis strains were grown in Luria-Bertani (LB, Difco, Detroit, MI) medium at 37 and 30oC, respectively. Where required, the growth medium was supplemented with chemical agents at the following concentrations: 2, 6-diaminopimelic acid (DAP), 30 µM; ampicillin, 50 µg/mL; kanamycin, 50 µg/mL; and gentamycin, 15 µg/mL.
Table 2. Strains and plasmids used in this study.
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| E. coli strain | ||
| DB3.1λ | Host for pMK2010 | 23 |
| WM3064 | Host for pir-dependent plasmids and donor strain for conjugation; ΔdapA | W. Metcalf, UIUC |
| S. oneidensis strains | ||
| MR-1 | Wild-type | Lab stock |
| HG0264 | As MR-1 plus ΔscyA | This study |
| HG0265 | As MR-1 plus ΔccmI | This study |
| HG0266 | As MR-1 plus ΔccmF (ccmF1) | This study |
| HG0268 | As MR-1 plus ΔccmH | This study |
| HG0269 | As MR-1 plus ΔSO0269 | This study |
| HG0478 | As MR-1 plus ΔccmF2 | This study |
| HG0478-6 | As MR-1 plus ΔSO0478 -6 | This study |
| HG1233 | As MR-1 plus ΔtorC | This study |
| HG1748 | As MR-1 plus ΔSO1748 | This study |
| HG2178 | As MR-1 plus ΔccpA | This study |
| HG3056 | As MR-1 plus ΔSO3056 | This study |
| Plasmids | ||
| pDS3.0 | Apr, Gmr, derivative from suicide vector pCVD442 | 24 |
| pMK2010 | Donor vector for ccdB-Camr cassette flanked by attP1 and attP2 | 23 |
| pHGM01 | Apr, Gmr, Cmr, att-based suicide vector | This study |
| pHG101 | Promoterless broad-host Kmr vector | 40 |
| pHG102 | pHG101 containing the S. oneidensis arcA promoter | 40 |
qRT-PCR
qRT-PCR analysis of RNA extracted from mid-logarithmic growing cells (OD600, ~0.4 and ~0.15 for aerobic and anaerobic cultures respectively) were carried out with an ABI7300 96-well qRT-PCR system (Applied Biosystems) essentially as described previously [37]. The expression of each gene was determined from three replicas in a single real-time qRT-PCR experiment. The Cycle threshold (C T) values for each gene of interest were averaged and normalized against the C T value of the arcA and recA genes, whose abundance was constant during the exponential phase [38,39]. Relative abundance (RA) of each gene was standardized to the C T values of both the arcA and recA genes using the equation RA = 2-∆CT, yielding similar fold differences.
Construction of in-frame mutants with a new mutagenesis-construct delivery plasmid employing BP recombination
The fusion PCR method used for construction of S. oneidensis in-frame deletion mutants involves with multiple rounds of PCR and conventional cloning, both of which greatly reduce its efficiency and effectiveness [24]. To make an improvement, in this study we developed a new mutagenesis-construct delivery vector employing attB-attP (BP) recombination based on the system reported before [24]. This plasmid, namely pHGM01, was created by introduction of a DNA fragment containing attP1, ccdB, CmR, and attP2 from pMK2010 into the SacI site of pDS3.0 [23,24] (Figure 2A). pHGM01 was propagated in E. coli strain DB3.1 λpir, which is able to maintain plasmids possessing a ccdB gene and an R6K γ origin of replication [23]. To construct an in-frame deletion mutant using pHGM01, attB1 and attB2 sequences were arranged next to gene specific sequences within the 5-O and 3-O primers, which located at each end of the fusion PCR product (in-frame deletion construct) for subsequent site specific recombination, which was performed using the BP Clonase (Invitrogen) according to the manufacturer’s instruction (Figure 2B). The resulting recombination mixture was transformed into E. coli strain WM3064, which permits the replication from the R6K γ origin but is sensitive to the ccdB gene. After recombination between attB and attP sequences, the fragment flanked by attP1 and attP2 within pHGM01 was replaced by the in-frame deletion construct, removing the ccdB gene. As a consequence, resultant destination vectors in which the recombination occurred enabled WM3064 cells to grow on plates containing ampicillin (and/or gentamycin) whereas the ccdB gene carried on the pHGM01 was toxic to WM3064. The correct destination vector for mutagenesis, verified by DNA sequencing, was then transferred into S. oneidensis by conjugation and the rest of the mutagenesis procedure was carried out the same as described before [24]. The final in-frame deletion mutant was confirmed by sequencing the mutated region.
Complementation of in-frame deletion mutants
Plasmids pHG101 and pHG102 were used in genetic complementation of mutants [40]. For complementation of genes next to their promoter, a fragment containing the gene of interest and its native promoter was generated by PCR and cloned into pHG101. For the rest genes, the gene of interest was amplified and inserted into MCS of pHG102 under the control of the arcA promoter, which is constitutively active [38]. Introduction of each verified complementation vector into the corresponding mutant was done by conjugation, and verified by PCR and restriction enzyme mapping.
Physiological characterization of mutant strains
For growth measurements under aerobic and anaerobic conditions, M1 defined medium containing 0.02% (w/v) of vitamin-free Casamino Acids was used as described previously [41,42]. Anaerobic media and cultures were prepared as reported earlier [18,41]. Growth of mutant strains was measured using a Bioscreen C microbiology reader (Labsystems Oy, Helsinki, Finland) by recording optical densities of cultures at 600 nm every 15 minutes. Generation times were calculated during the exponential growth phase.
Biochemical methods
Cells of the mid-exponential phase were harvested and then were lysed with lysis buffer (0.25 M Tris/HCl, (pH 7.5), 0.5% Trion-X100). Protein concentration was determined with a bicinchoninic acid assay kit with bovine serum albumin (BSA) as a standard according to the manufacturer’s instructions (Pierce Chemical). The amount of heme c was measured following the procedure described elsewhere [43].
Statistical analyses
Statistical significance of the difference between experimental groups was assessed by two-way analysis of variance (ANOVA) followed by Bonferroni posttests.
Acknowledgments
We thank Michael L. Kahn (Washington State U. USA) for E. coli DB3.1 λpir and plasmid pMK2010 that served as foundation for the plasmid constructions and recombination procedures described in this work.
Funding Statement
This research was supported by Major State Basic Research Development Program (973 Program: 2010CB833803), National Natural Science Foundation of China (31270097), and Natural Science Foundation of Zhejiang province (R3110096) to HG. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Kranz RG, Richard-Fogal C, Taylor J-S, Frawley ER (2009) Cytochrome c biogenesis: Mechanisms for covalent modifications and trafficking of heme and for heme-iron redox control. Microbiol Mol Biol Rev 73: 510-528. doi:10.1128/MMBR.00001-09. PubMed: 19721088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sanders C, Turkarslan S, Lee D-W, Daldal F (2010) Cytochrome c biogenesis: the Ccm system. Trends Microbiol 18: 266-274. doi:10.1016/j.tim.2010.03.006. PubMed: 20382024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stevens JM, Mavridou DAI, Hamer R, Kritsiligkou P, Goddard AD et al. (2011) Cytochrome c biogenesis System I. FEBS J 278: 4170-4178. doi:10.1111/j.1742-4658.2011.08376.x. PubMed: 21958041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Simon J, Eichler R, Pisa R, Biel S, Gross R (2002) Modification of heme c binding motifs in the small subunit (NrfH) of the Wolinella succinogenes cytochrome c nitrite reductase complex. FEBS Lett 522: 83-87. doi:10.1016/S0014-5793(02)02885-5. PubMed: 12095623. [DOI] [PubMed] [Google Scholar]
- 5. Fülöp V, Sam KA, Ferguson SJ, Ginger ML, Allen JWA (2009) Structure of a trypanosomatid mitochondrial cytochrome c with heme attached via only one thioether bond and implications for the substrate recognition requirements of heme lyase. FEBS J 276: 2822-2832. doi:10.1111/j.1742-4658.2009.07005.x. PubMed: 19459937. [DOI] [PubMed] [Google Scholar]
- 6. Heidelberg JF, Paulsen IT, Nelson KE, Gaidos EJ, Nelson WC et al. (2002) Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis . Nat Biotechnol 20: 1118-1123. doi:10.1038/nbt749. PubMed: 12368813. [DOI] [PubMed] [Google Scholar]
- 7. Fredrickson JK, Romine MF, Beliaev AS, Auchtung JM, Driscoll ME et al. (2008) Towards environmental systems biology of Shewanella . Nat Rev Microbiol 6: 592-603. doi:10.1038/nrmicro1947. PubMed: 18604222. [DOI] [PubMed] [Google Scholar]
- 8. Meyer TE, Tsapin AI, Vandenberghe I, De Smet L, Frishman D et al. (2004) Identification of 42 possible cytochrome c genes in the Shewanella oneidensis genome and characterization of six soluble cytochromes. Omics 8: 57-77. doi:10.1089/153623104773547499. PubMed: 15107237. [DOI] [PubMed] [Google Scholar]
- 9. Romine MF, Carlson TS, Norbeck AD, McCue LA, Lipton MS (2008) Identification of mobile elements and pseudogenes in the Shewanella oneidensis MR-1 genome. Appl Environ Microbiol 74: 3257-3265. doi:10.1128/AEM.02720-07. PubMed: 18378659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gao H, Barua S, Liang Y, Wu L, Dong Y et al. (2010) Impacts of Shewanella oneidensis c-type cytochromes on aerobic and anaerobic respiration. J Microbiol Biotechnol 3: 455-466. doi:10.1111/j.1751-7915.2010.00181.x. PubMed: 21255343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mowat CG, Rothery E, Miles CS, McIver L, Doherty MK et al. (2004) Octaheme tetrathionate reductase is a respiratory enzyme with novel heme ligation. Nat Struct Mol Biol 11: 1023-1024. doi:10.1038/nsmb827. PubMed: 15361860. [DOI] [PubMed] [Google Scholar]
- 12. Atkinson SJ, Mowat CG, Reid GA, Chapman SK (2007) An octaheme c-type cytochrome from Shewanella oneidensis can reduce nitrite and hydroxylamine. FEBS Lett 581: 3805-3808. doi:10.1016/j.febslet.2007.07.005. PubMed: 17659281. [DOI] [PubMed] [Google Scholar]
- 13. Hartshorne RS, Kern M, Meyer B, Clarke TA, Karas M et al. (2007) A dedicated haem lyase is required for the maturation of a novel bacterial cytochrome c with unconventional covalent haem binding. Mol Microbiol 64: 1049-1060. doi:10.1111/j.1365-2958.2007.05712.x. PubMed: 17501927. [DOI] [PubMed] [Google Scholar]
- 14. Lezhneva L, Kuras R, Ephritikhine G, de Vitry C (2008) A novel pathway of cytochrome c biogenesis is involved in the assembly of the cytochrome b 6f complex in Arabidopsis chloroplasts. J Biol Chem 283: 24608-24616. doi:10.1074/jbc.M803869200. PubMed: 18593701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cianciotto NP, Cornelis P, Baysse C (2005) Impact of the bacterial type I cytochrome c maturation system on different biological processes. Mol Microbiol 56: 1408-1415. doi:10.1111/j.1365-2958.2005.04650.x. PubMed: 15916594. [DOI] [PubMed] [Google Scholar]
- 16. Richter K, Schicklberger M, Gescher J (2012) Dissimilatory reduction of extracellular electron acceptors in anaerobic respiration. Appl Environ Microbiol 78: 913-921. doi:10.1128/AEM.06803-11. PubMed: 22179232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nissen S, Liu X, Chourey K, Hettich RL, Wagner DD, Pfiffner SM et al. (2012) Comparative c-type cytochrome expression analysis in Shewanella oneidensis strain MR-1 and Anaeromyxobacter dehalogenans strain 2CP-C grown with soluble and insoluble oxidized metal electron acceptors. Biochem Soc Trans 40(6): 1204-1210. doi:10.1042/BST20120182. PubMed: 23176455. [DOI] [PubMed] [Google Scholar]
- 18. Gao H, Yang ZK, Barua S, Reed SB, Romine MF et al. (2009) Reduction of nitrate in Shewanella oneidensis depends on atypical NAP and NRF systems with NapB as a preferred electron transport protein from CymA to NapA. ISME J 3: 966-976. doi:10.1038/ismej.2009.40. PubMed: 19387485. [DOI] [PubMed] [Google Scholar]
- 19. Beliaev AS, Klingeman DM, Klappenbach JA, Wu L, Romine MF et al. (2005) Global transcriptome analysis of Shewanella oneidensis MR-1 exposed to different terminal electron acceptors. J Bacteriol 187: 7138-7145. doi:10.1128/JB.187.20.7138-7145.2005. PubMed: 16199584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schwalb C, Chapman SK, Reid GA (2003) The tetraheme cytochrome CymA is required for anaerobic respiration with dimethyl sulfoxide and nitrite in Shewanella oneidensis . Biochemistry 42: 9491-9497. doi:10.1021/bi034456f. PubMed: 12899636. [DOI] [PubMed] [Google Scholar]
- 21. McMillan DGG, Marritt SJ, Butt JN, Jeuken LJC (2012) Menaquinone-7 is specific cofactor in tetraheme quinol dehydrogenase CymA. J Biol Chem 287: 14215-14225. doi:10.1074/jbc.M112.348813. PubMed: 22393052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhou G, Yin J, Chen H, Hua Y, Sun L et al. (2013) Combined effect of loss of the caa 3 oxidase and Crp regulation drives Shewanella to thrive in redox-stratified environments. ISME J, 7: 1752–63. doi:10.1038/ismej.2013.62. PubMed: 23575370. PubMed: 23575370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. House BL, Mortimer MW, Kahn ML (2004) New recombination methods for Sinorhizobium meliloti genetics. Appl Environ Microbiol 70: 2806-2815. doi:10.1128/AEM.70.5.2806-2815.2004. PubMed: 15128536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gao W, Liu Y, Giometti CS, Tollaksen SL, Khare T et al. (2006) Knock-out of SO1377 gene, which encodes the member of a conserved hypothetical bacterial protein family COG2268, results in alteration of iron metabolism, increased spontaneous mutation and hydrogen peroxide sensitivity in Shewanella oneidensis MR-1. BMC Genomics 7: 76. doi:10.1186/1471-2164-7-76. PubMed: 16600046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gon S, Patte J-C, Dos Santos J-P, Méjean V (2002) Reconstitution of the trimethylamine oxide reductase regulatory elements of Shewanella oneidensis in Escherichia coli . J Bacteriol 184: 1262-1269. doi:10.1128/JB.184.5.1262-1269.2002. PubMed: 11844754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Saltikov CW, Newman DK (2003) Genetic identification of a respiratory arsenate reductase. Proc Natl Acad Sci U S A 100: 10983-10988. doi:10.1073/pnas.1834303100. PubMed: 12939408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Thormann KM, Saville RM, Shukla S, Spormann AM (2005) Induction of rapid detachment in Shewanella oneidensis MR-1 biofilms. J Bacteriol 187: 1014-1021. doi:10.1128/JB.187.3.1014-1021.2005. PubMed: 15659679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Eaves DJ, Grove J, Staudenmann W, James P, Poole RK et al. (1998) Involvement of products of the nrfEFG genes in the covalent attachment of haem c to a novel cysteine–lysine motif in the cytochrome c 552 nitrite reductase from Escherichia coli . Mol Microbiol 28: 205-216. PubMed: 9593308. [DOI] [PubMed] [Google Scholar]
- 29. Pisa R, Stein T, Eichler R, Gross R, Simon J (2002) The nrfI gene is essential for the attachment of the active site haem group of Wolinella succinogenes cytochrome c nitrite reductase. Mol Microbiol 43: 763-770. doi:10.1046/j.1365-2958.2002.02784.x. PubMed: 11929530. [DOI] [PubMed] [Google Scholar]
- 30. Einsle O (2011) Structure and function of formate-dependent cytochrome c nitrite reductase, NrfA. In: Martin GK, Lisa YS. Methods in Enzymology. Academic Press; pp. 399-422. [DOI] [PubMed] [Google Scholar]
- 31. Shirodkar S, Reed S, Romine M, Saffarini D (2011) The octahaem SirA catalyses dissimilatory sulfite reduction in Shewanella oneidensis MR-1. Environ Microbiol 13: 108-115. doi:10.1111/j.1462-2920.2010.02313.x. PubMed: 21199252. [DOI] [PubMed] [Google Scholar]
- 32. Fonseca BM, Paquete CM, Neto SE, Pacheco I, Soares CM et al. (2013) Mind the gap: cytochrome interactions reveal electron pathways across the periplasm of Shewanella oneidensis MR-1. Biochem J 449: 101-108. doi:10.1042/BJ20121467. PubMed: 23067389. [DOI] [PubMed] [Google Scholar]
- 33. Schütz B, Seidel J, Sturm G, Einsle O, Gescher J (2011) Investigation of the electron transport chain to and the catalytic activity of the diheme cytochrome c peroxidase CcpA of Shewanella oneidensis . Appl Environ Microbiol 77: 6172-6180. doi:10.1128/AEM.00606-11. PubMed: 21742904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Londer YY, Giuliani SE, Peppler T, Collart FR (2008) Addressing Shewanella oneidensis “cytochromome”: The first step towards high-throughput expression of cytochromes c . Protein Expr Purif 62: 128-137. doi:10.1016/j.pep.2008.06.014. PubMed: 18657620. [DOI] [PubMed] [Google Scholar]
- 35. Bretschger O, Obraztsova A, Sturm CA, Chang IS, Gorby YA et al. (2007) Current production and metal oxide reduction by Shewanella oneidensis MR-1 wild type and mutants. Appl Environ Microbiol 73: 7003-7012. doi:10.1128/AEM.01087-07. PubMed: 17644630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lukat P, Rudolf M, Stach P, Messerschmidt A, Kroneck PMH et al. (2008) Binding and reduction of sulfite by cytochrome c nitrite reductase. Biochemistry 47: 2080-2086. doi:10.1021/bi7021415. PubMed: 18201106. [DOI] [PubMed] [Google Scholar]
- 37. Yuan J, Wei B, Shi M, Gao H (2011) Functional assessment of EnvZ/OmpR two-component system in Shewanella oneidensis . PLOS ONE 6: e23701. doi:10.1371/journal.pone.0023701. PubMed: 21886811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gao H, Wang X, Yang ZK, Chen J, Liang Y et al. (2010) Physiological roles of ArcA, Crp, and EtrA and their interactive control on aerobic and anaerobic respiration in Shewanella oneidensis . PLOS ONE 5: e15295. doi:10.1371/journal.pone.0015295. PubMed: 21203399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gralnick JA, Vali H, Lies DP, Newman DK (2006) Extracellular respiration of dimethyl sulfoxide by Shewanella oneidensis strain MR-1. Proc Natl Acad Sci U S A 103: 4669-4674. doi:10.1073/pnas.0505959103. PubMed: 16537430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wu L, Wang J, Tang P, Chen H, Gao H (2011) Genetic and molecular characterization of flagellar assembly in Shewanella oneidensis . PLOS ONE 6: e21479. doi:10.1371/journal.pone.0021479. PubMed: 21731763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gao H, Wang X, Yang ZK, Palzkill T, Zhou J (2008) Probing regulon of ArcA in Shewanella oneidensis MR-1 by integrated genomic analyses. BMC Genomics 9: 42. doi:10.1186/1471-2164-9-42. PubMed: 18221523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Myers CR, Nealson KH (1990) Respiration-linked proton translocation coupled to anaerobic reduction of manganese(IV) and iron(III) in Shewanella putrefaciens MR-1. J Bacteriol 172: 6232-6238. PubMed: 2172208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Berry EA, Trumpower BL (1987) Simultaneous determination of hemes a, b, and c from pyridine hemochrome spectra. Anal Biochem 161: 1-15. doi:10.1016/0003-2697(87)90643-9. PubMed: 3578775. [DOI] [PubMed] [Google Scholar]