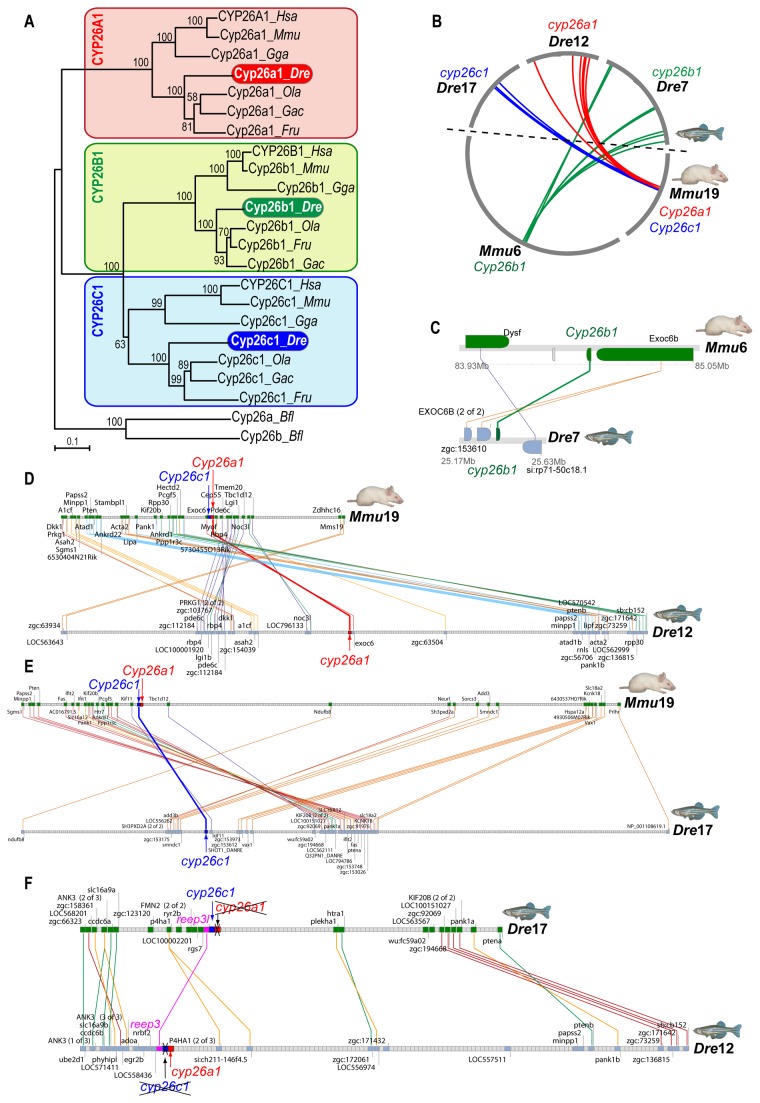
Figure 2. Evolutionary relationships of CYP26 family members in zebrafish and mouse.
(A) Phylogenetic analyses inferred by maximum likelihood (ML) indicate that Cyp26 paralogs of teleosts (i.e. zebrafish (Dre), medaka (Ola), stickleback (Gac) and fugu (Tru)) grouped with their correspondingly named Cyp26 paralogs of tetrapods (i.e. mouse (Mmu), human (Hsa) and chicken (Gga)). Numbers indicate bootstrap values supporting each node (n=100), and no significant differences were found between ML and NJ analyses. (B) A circleplot shows graphically orthologous relationships of cyp26 genomic neighborhoods shared between zebrafish and mouse. Grey circumferential arcs represent chromosomes, with Cyp26b1 in green on Mmu6, Cyp26a1 in red on Mmu19, and Cyp26c1 in blue on Mmu19. Colored lines link orthologous regions in zebrafish chromosomes Dre7, Dre12 and Dre17, the sites of cyp26b1, cyp26a1 and cyp26c1 genes, respectively. (C–E) Clusters of synteny conservation reveal the presence of many gene neighbor orthologs shared between each Cyp26 genomic neighborhood in mouse and zebrafish cyp26b1 (C: cluster ID#265419 according to the Synteny Database), cyp26a1 (D: cluster ID#258723) and cyp26c1 (E: cluster ID#265367). These results rule out the possibility of reciprocal gene losses in zebrafish and mouse that could mask actual orthologous relationships in artifactual phylogenetic trees, and provide strong support for the conclusion that the zebrafish/mouse gene pairs called cyp26a1/Cyp26a1 and cyp26b1/Cyp26b1 are in fact orthologs. F: Gene clusters of synteny conservation (cluster ID#191383) between Dre12 and Dre17 reveal that the genomic neighborhoods of cyp26a1 and cyp26c1 are paralogous due to the teleost genome duplication (TGD, R3) that preceded the divergence of the teleost lineage after it split from the tetrapod branch. Duplicates of cyp26a1 and cyp26c1 in Dre17 and Dre12 were lost reciprocally after the teleost genome duplication (labeled with crosses) in contrast to, for instance, reep3 paralogs (in pink) that were maintained adjacent to cyp26 paralogs.