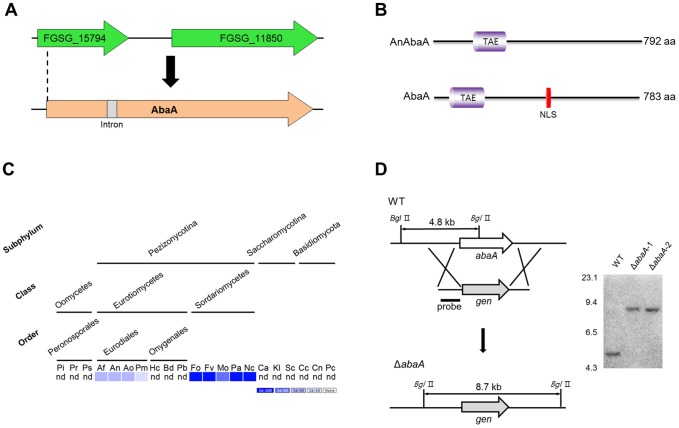
Figure 1. Identification and deletion of abaA in F. graminearum.
(A) Re-annotation of the abaA ortholog gene in F. graminearum. cDNA sequencing analysis revealed an exact abaA open reading frame with an intron. (B) Comparison of AbaA orthologs between A. nidulans and F. graminearum. A schematic representation of the protein orthologs shows the location and alignment of conserved domains. The TAE and TEA/ATTS domains (IPR000818) predicted with InterProScan; NLS, nuclear localization signal predicted by using NLStradamus [62]. (C) Distribution of AbaA homologs in representative fungal species. The distribution image was constructed using the BLASTMatrix tool that is available on the Comparative Fungal Genomics Platform (http://cfgp.riceblast.snu.ac.kr/) [74]. Pi, Phytophthora infestans; Pr, P. ramorum; Ps, P. sojae; Af, Aspergillus fumigatus; An, A. nidulans; Ao, A. oryzae; Pm, Penicillium marneffei, Hc, Histoplasma capsulatum; Bd, Blastomyces dermatitidis; Pb, Paracoccidioides brasiliensis; Fo, Fusarium oxysporum; Fv, F. verticillioides; Mo, Magnaporthe oryzae; Pa, Podospora anserine; Nc, Neurospora crassa; Ca, Candida albicans; Kl, Kluyveromyces lactis; Sc, Saccharomyces cerevisiae; Cc, Coprinus cinereus; Cn, Cryptococcus neoformans; Pc, Phanerochaete chrysosporium; nd, not detected. (D) Targeted deletion of the abaA gene. The abaA gene was deleted from the F. graminearum wild-type genome. Left panel, schematic representation of the homologous gene recombination strategy used to generate the abaA deletion mutants. Right panel, Southern blot analysis. Sizes of the DNA standards used are indicated in kilobases to the left of the blot.