Abstract
Considerable Nanoarchaeota novelty and diversity were encountered in Yellowstone Lake, Yellowstone National Park (YNP), where sampling targeted lake floor hydrothermal vent fluids, streamers and sediments associated with these vents, and in planktonic photic zones in three different regions of the lake. Significant homonucleotide repeats (HR) were observed in pyrosequence reads and in near full-length Sanger sequences, averaging 112 HR per 1349 bp clone and could confound diversity estimates derived from pyrosequencing, resulting in false nucleotide insertions or deletions (indels). However, Sanger sequencing of two different sets of PCR clones (110 bp, 1349 bp) demonstrated that at least some of these indels are real. The majority of the Nanoarchaeota PCR amplicons were vent associated; however, curiously, one relatively small Nanoarchaeota OTU (71 pyrosequencing reads) was only found in photic zone water samples obtained from a region of the lake furthest removed from the hydrothermal regions of the lake. Extensive pyrosequencing failed to demonstrate the presence of an Ignicoccus lineage in this lake, suggesting the Nanoarchaeota in this environment are associated with novel Archaea hosts. Defined phylogroups based on near full-length PCR clones document the significant Nanoarchaeota 16S rRNA gene diversity in this lake and firmly establish a terrestrial clade distinct from the marine Nanoarcheota as well as from other geographical locations.
Keywords: Nanoarchaeota, Yellowstone Lake, pyrosequencing
Introduction
Huber et al. (2002) described the cultivation of a novel hyperthermophilic archeaon coined Nanoarchaeum equitans. This organism requires the host organism Ignicoccus hospitalis, living as an obligate parasite because it lacks genes coding for biosynthesis of essential cellular components such as lipids, cofactors, amino acids, or nucleotides (Waters et al., 2003). Thus far, N. equitans is the lone cultured representative of the archaeal subdivision Nanoarchaeota, though PCR-based environmental studies have found the Nanoarchaeota in several high temperature marine environments (Hohn et al., 2002; Stetter et al., 2005; McCliment et al., 2006; Roussel et al., 2011; Flores et al., 2011, 2012). In addition, the Nanoarchaeota 16S signature has been documented in hypersaline mats (Casanueva et al., 2008), suggesting these organisms are more widely distributed than in geo/hydrothermal environments.
Yellowstone National Park (YNP) is a well-known high temperature environment that has been extensively studied, in particular during the last decade. In addition to being home to a wide range of organisms belonging to the domain Bacteria (e.g., Hugenholtz et al., 1998; Fouke et al., 2000; Reysenbach et al., 2000; Jackson et al., 2001; Botero et al., 2005; Spear et al., 2005; Yang et al., 2011), documentation of the Archaea in the YNP geothermal microbial communities has also been accumulating (e.g., Barns et al., 1994; Karavaĭko et al., 1994; Auchtung et al., 2006; Boyd et al., 2007; Ellis et al., 2008; Inskeep et al., 2010; Kan et al., 2011). Thus, far, three reports of the Nanoarchaeota have been noted for YNP, with all involving work at Obsidian Pool. Hohn et al. (2002) PCR cloned Nanoarchaeota 16S rRNA genes from Obsidian Pool, referred to as clone OP9. Later, Stetter et al. (2005) examined Obsidian Pool samples with Nanoarchaeota-specific FISH probes and described “tiny cocci, about the size of N. equitans attached to the surface of Pyrobaculum-shaped rods that may represent these novel nanoarchaeotes.” And just recently, Podar et al. (2013) used cell sorting techniques to capture and genome sequence Nanoarchaeota cells (coined Nst1) and their inferred hosts from Obsidian Pool, YNP. This latter study represents an important advancement in Nanoarchaeota biology in that the genome comparison with that of N. equitans revealed clear differences (e.g., less reduction). Importantly, Podar et al. (2013) also provided evidence of the Nanoarchaeota associated with host Archaea different from Ignicoccus, inferred to be a Sulfolobales-like organism in this case (Podar et al., 2013).
While these contemporary efforts have made foundational changes to our understanding of the microbial diversity and distribution in the YNP geothermal complex, many geotherm features remain to be studied. One such environment is Yellowstone Lake. It is the highest elevation (~2300 m) among large subalpine high-altitude lakes in North America (Morgan et al., 2007), with a maximum measured depth of 131 m and average depth of 42.5 m (Benson, 1961; Morgan et al., 2007). Several studies conducted by the United States Geologic Survey (USGS) have documented extinct or active hydrothermal vents at specific locations on the lake floor (Morgan et al., 2003; Balistrieri et al., 2007; Morgan et al., 2007). Thus, in addition to the microbiota that normally comprise the microbial community of a sub-alpine lake, there are substantial opportunities to study thermophiles associated with the lake floor vents.
We have recently conducted extensive surveys of this lake (Clingenpeel et al., 2011; Kan et al., 2011), characterizing its high-energy geochemistry and substantial microbial diversity, and documenting microbial phylotypes previously known to only occur in marine environments. Here we describe additional work with the vents and photic zones in this lake, though in this case targeting the Nanoarchaeota. We describe the geochemistry and summarize the results from pyrosequencing and Sanger sequencing of 16S rRNA gene clones generated with Nanoarchaeota specific PCR primers. The cloned sequences document a North American clade of the Nanoarchaeota and the significant diversity therein.
Materials and methods
Lake locations and sampling
Lake sampling took place in September 2007 and 2008. Vent fields were located based on global information system coordinates established from past USGS surveys (e.g., Morgan et al., 1977, 2007). Individual vents were located and sampled by remote operated vehicle (ROV) reconnaissance of the lake floor within the Inflated Plain and West Thumb regions of the lake. Lake location and brief description of each site and of within-site samples are described below, and the relative and approximate lake locations are shown in Figure 1. Vent fluids and streamer samples were collected using a boat-tethered ROV previously described (Lovalvo et al., 2010; Clingenpeel et al., 2011; Kan et al., 2011). Characterization for aqueous solutes and gases were as recently described (Clingenpeel et al., 2011).
Figure 1.
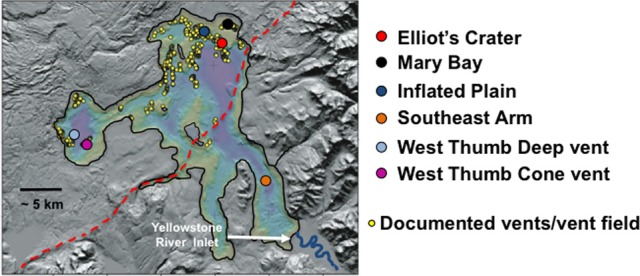
A relief map illustrating Yellowstone Lake and the approximate sampling locations described in this study. Image is a modification from that published in Morgan et al. (2007) and is used here with permission. Red dashed line represents the approximate boundary of the Yellowstone caldera. Relief map image kindly provided by Lisa Morgan.
Biomass sampling methods were as described by Clingenpeel et al. (2011). Briefly, 100–300 l of lake or vent water was pumped through a 20 μm pre-filter into 50 l carboys on the boat deck. Carboys were sterilized prior to use by autoclaving or by bleaching followed by rinsing with autoclaved distilled water. Using techniques previously described for the global ocean survey (Rusch et al., 2007), the biomass in the lake and vent water was size fractionated by serial filtration through 3.0, 0.8, and 0.1 μm membrane filters. Filters were sealed in plastic bags and frozen for transport to the laboratory at Montana State University, where they were stored at −80°C. To obtain microbial streamer samples from the vent flow stream, the ROV sampling arm was positioned over the streamer structure and then an ROV vacuum device was engaged to suction the streamer biomass into a holding canister on the ROV (see Lovalvo et al., 2010).
Nucleic acid extraction, PCR, and sequencing
DNA was extracted as described by Clingenpeel et al. (2011). Full-length 16S rRNA gene amplification for clone library construction and generation of shorter amplicons for pyrosequencing were performed using primers described in Table 1. Near full-length PCR products were cloned using the TOPO TA Cloning Kit (Invitrogen Corp.), with inserts sequenced using the Big Dye Terminator chemistry (Applied Biosystems) and the Applied Biosystems 3100 Genetic Analyzer. Chimeric sequences were screened by the “CHIMERA DETECTION” program of the Ribosomal Database Project Maidak et al. (1997) and removed from further analysis.
Table 1.
Primers used in this study.
| Primer | Sequence (5′–3′) | Target groups | References |
|---|---|---|---|
| FULL-LENGTH CLONING | |||
| N3F | TCCCGTTGATCCTGCG | Nanoarchaeota | Huber et al., 2002 |
| N1406R | ACGGGCGGTGAGTGCAA | Nanoarchaeota | Huber et al., 2002 |
| YNP 35F | TCCCTCCGACTAACCCATGG | YNP Nanoarchaeota | This study |
| YNP 1337R | ACCGGGGGAATAGTGACC | YNP Nanoarchaeota | This study |
| PRIMERS FOR PYRO SEQUENCING | |||
| N3aF* | TCCCGTTGATCCTGCGG | Nanoarchaeota | This study |
| N3bF* | TCCAGTTGATCCTGCGGG | Nanoarchaeota | This study |
| N3cF* | TCCCGTGTGATCCTGCG | Nanoarchaeota | This study |
| N495R | TGGCGACTGCCACCCCT | Nanoarchaeota | This study |
Modified from N3F.
The V1 + V2 region of the 16S rRNA gene was amplified for 454 Titanium pyrosequencing using primers developed in this study (Table 1). After 25 cycles of amplification, 5 more cycles were used to add the sample specific barcodes and the adaptor sequences required for 454 pyrosequencing. The barcoded 16S rRNA gene PCR amplicons obtained from the different environments were pooled, with the volume of each sample qualitatively adjusted to reflect the strength of the amplicon.
Near full-length clone libraries were aligned, trimmed, and then initially classified using BLAST (Altschul et al., 1990), and can be found as GenBank accession numbers JF262403–JF262535. Neighbor-Joining distance trees were constructed using MacVector 10.0 software package (GCG) and Maximum Likelihood trees were constructed using PhyML web interface (http://www.atgc-montpellier.fr/phyml/). In both analyses, bootstraps were generated from 1000 resampling datasets. OTU groupings were assigned using ARB software (Ludwig et al., 2004) and the latest released Silva 102 database (Pruesse et al., 2007). The pyrosequencing reads were quality trimmed according to Kunin et al. (2010) followed by clustering using abundance-sorted preclustering per Huse et al. (2010) and a final complete linkage (furthest neighbor) clustering using the mothur software (Schloss et al., 2009). Collector's curve analysis was also done in mothur. The identification of pyrosequencing reads as nanoarchaea was done by classification with the RDP Classifier (Wang et al., 2007; Cole et al., 2009). Techniques we previously described (Clingenpeel et al., 2011; Kan et al., 2011) were used to match the pyrosequencing reads with the near full-length Sanger sequenced clones. Briefly, the pyroreads were compared to the near full-length clone sequences using BLAST (Altschul et al., 1990), with match criteria requiring ≥99% identity for ≥95% of the read length in order to be assigned to a phylogroup. The pyroreads can be found under the identifiers SRS150246 and SRS150227 in the SRA database.
Results
Sampling sites and geochemistry
The lake sampling sites examined in this study are shown in Figure 1. Two sampling sites were located in the West Thumb region and three sites in the northern portion of the lake, referred to as Elliot's Crater (a lake floor geologic feature), Mary Bay, and Inflated Plain. All of these locations correspond to lake floor vent fields previously documented by (Morgan et al., 2003, 2007) (Figure 1) and on which we have reported on recently (Lovalvo et al., 2010; Clingenpeel et al., 2011; Kan et al., 2011). As a control environment to contrast with the vent field regions of the lake, one site in the Southeast Arm was also included. This location is well-outside the caldera boundary, not known to be associated with any vent activity (Morgan et al., 2003, 2007), and most proximal to the primary tributary to the lake, the Yellowstone River. Prior to flowing into the Southeast Arm, the Yellowstone River does not drain geothermal features elsewhere in YNP (YNP Ground Surveys, Spatial Analysis Center 2005; Savage et al., 2012).
Geochemical parameters of significance to microbial selection are summarized in Table 2. Vent emissions varied in pH (5.2–6.7) and temperature (37–73°C), with the latter also exhibiting within-vent variation documented as temperature surges determined by real time ROV monitoring during sample acquisition. Gas composition varied between and within vent fields (Table 2). As examples, vent H2 levels in the northern regions of the lake (Inflated Plain, Mary Bay, Elliot's Crater) were consistently much greater than in the West Thumb vents (Table 2). The photic zone water chemistry varied, depending on whether the samples were acquired in water columns overlying lake floor vents and in such cases reflected the constituents observed in the vent emissions located directly below. For instance, in 10 m photic zone samples associated with the high output Inflated Plain vents (Table 2), levels of CH4, H2, and CO2 were orders of magnitude higher and pH more acidic (6.1–6.6) than the Southeast Arm photic waters (Table 2), which were neutral pH, cold and well-aerated. It is worth noting that NH4, CO2, CH4, and H2 in the Southeast Arm were still at microbially relevant concentrations.
Table 2.
Sample identification, lake location, and general characteristics. Some geochemical parameters were determined in duplicate.
| Lake ID no. | Pyrosequencing reads | Lake location and sample type | Temp.(C°) | pH | Depth (m) | Selected nutrients and energy sources | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| NH4 | CO2(aq) | S2−(aq) | O2(aq) | CH4(aq) | H2(aq) | ||||||
| uM | mM | uM | uM | uM | uM | ||||||
| ELLIOT'S CRATER | |||||||||||
| 1 | 1672 | Elliot's Crater Vent emissions | 63–68 | 6.4 | 14.1 | 45.0 | 0.49,47 | 21.7 | 119 | 2.5, 2.1 | 762, 558 |
| MARY BAY | |||||||||||
| 2 | 2186 | Vent steamers and sediments | 62–82 | 5.2 | 50.5 | 53.8 | 3.77, 1.80 | 79.5 | bd | 28.1, 12.4 | 2984, 2797 |
| INFLATED PLAIN | |||||||||||
| 3 | 257 | IP photic 10 m 0.1 um filter | 12.2 | 6.1 | 10 | 2.6 | 0.11, 0.11, 0.10 | 2.5 | 261 | 2.4, 2.7, 2.6 | 773, 798, 716 |
| 4 | 1553 | IP photic 10 m 0.8 um filter | |||||||||
| 5 | 252 | IP photic 10 m 3.0 um filter | |||||||||
| 6 | 3221 | IP vent steamer 1 | 40–60 | 5.2 | 30 | 30.9 | 3.1, 3.2 | 248 | bd++ | 20.9, 22.5 | 1031, 1430 |
| 7 | 2510 | IP steamer 2 | |||||||||
| 8 | 2222 | IP vent 2 emissions | 44–52 | 5.6 | 33.6 | 8.2 | 1.1, 1.1 | 98 | bd | 6.7, 5.4 | 1974 |
| 9 | 1996 | IP mixing zone water | 21–30 | 5.6 | 33.3 | 37.5 | 0.57 | 118 | 23 | 2.7 | 386 |
| WEST THUMB | |||||||||||
| 10 | 3309 | WT deep vent emissions (2007) | 60–66 | 6.2 | 52.0 | 12.1 | 1.7 | 2.1 | 113 | 6.4 | 49 |
| 11 | 2797 | WT deep vent emissions (2008) | 40–73 | 6.0 | 52.3 | 23.4 | 2.4 | 11.0 | 197 | 7.6 | 32 |
| 12 | 1959 | WT deep vent steamer | 40–73 | 6.0 | 52.3 | 23.4 | 2.4 | 11.0 | 197 | 7.6 | 32 |
| 13 | 1904 | WT deep vent mixing zone water | 26 | 6.6 | 52.1 | 27.2 | ND* | 0.3 | 211 | ND | ND |
| 14 | 638 | West thumb cone vent | 37 | 6.7 | 26.1 | 27.4 | 10.22 | bd | 82 | 7.6 | 14 |
| SOUTHEAST ARM | |||||||||||
| 15 | 377 | SEA photic 10 m 0.1 um filter | 12.3 | 7.0 | 10.0 | 6.2 | 0.02 | 0.1 | 273 | 0.1 | 33 |
| 16 | 1221 | SEA photic 10 m 0.8 um filter | |||||||||
| 17 | 412 | SEA photic 10 m 3.0 um filter | |||||||||
ND, Not determined.
bd, Below detection.
Nanoarchaeote diversity: near full-length sanger sequencing
A total of 131 near full-length Nanoarchaeota 16S rDNA PCR clones were obtained from vent or photic zone water samples. Sanger sequencing revealed considerable within-lake diversity (Figure 2). All lake clones were most closely related to the YNP Nanoarchaeota clones derived from Obsidian Pool (Hohn et al., 2002) located ~8 km from the lake. Further, all YNP clones branched distinctly separate from N. equitans and from the Kamchatka environmental clones (Figure 2). These near full-length clones were grouped based on bootstrap-supported branching cluster relatedness and designated as clone groups A–F (Figure 2). When examined using the neighbor-joining algorithm, phylogroups C and D are separate clades and phylogroup F is a single group. However, when examined using maximum likelihood, phylogroups C and D merge and phylogroup F splits into two smaller clusters (Figure 2). Consensus sequences were generated for each phylogroup (nucleotide assignments based on majority rule) and then compared and used to generate a lake-wide consensus sequence. Comparing the phylogroup consensus sequences against the lake-wide consensus sequence illustrated a total of 136 points of sequence divergence scattered across the cloned region, but with ~55% of the diversity occurring in the 550–850 nt region of the near full length clones. There were many instances of insertions and deletions (indels) observed in these comparisons.
Figure 2.
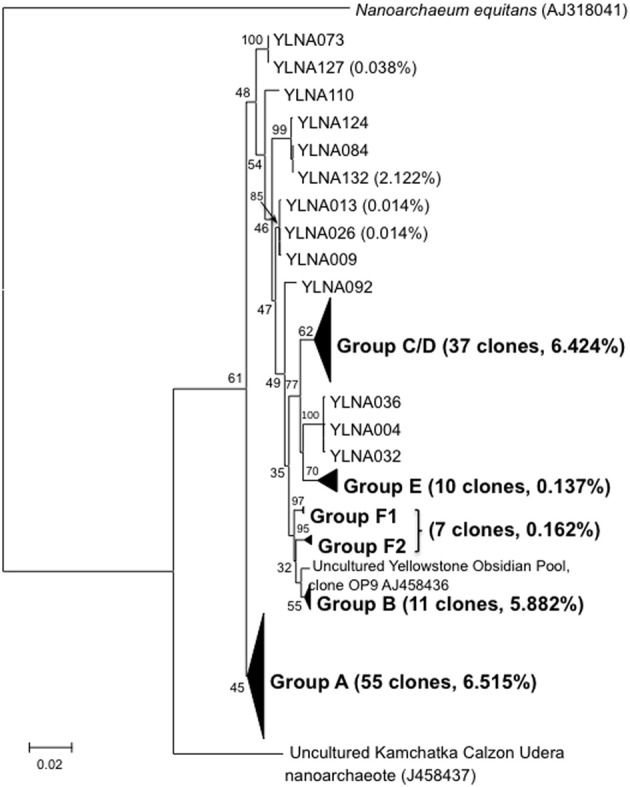
Maximum likelihood tree illustrating the phylogenetic relatedness of the Yellowstone like near full-length Nanoarchaeota clones relative to other near full-length clones and to N. queans. For each clone group, the number of near full-length clones obtained in this study and the percentage of proofreads that match the Sanger Sequence is provided in parentheses. Only relevant bootstrap values are shown.
Nanoarchaeote diversity: pyrosequencing
The PCR amplicons from the different samples varied in strength, with the strongest amplicons deriving from samples associated with vents (fluid emissions, streamers, or mixing zones where vent fluids mixed with lake water). After quality trimming, 28,441 pyrosequencing reads were advanced to further analysis. Read number for each sample (Table 2) reflects the amplicon strength for each sample that then determined how much of each were pooled for the barcoded pyrosequencing effort. The resulting sequence microdiversity was significant, again with a high frequency of apparent indels. To further investigate these indels, additional PCRs were conducted to individually clone and Sanger sequence a 113 bp region of the Nanoarchaeota 16S gene (positions 365–478 in the N. equitans 16S gene, primarily the conserved region between V2 and V3). From a small sample (31 clones), many of the indels found in the pyrosequencing reads could be identically matched with the Sanger sequenced longer clones (results not shown). Since homonucleotide repeats (HRs) may contribute to these indels in pyrosequenced DNA, the near full-length Sanger-sequenced clones were examined in more detail for this feature. There were 112 ± 4 HRs of 3–7 bp in length occurring across the near full-length clones. Further analysis of these near full-length clones identified 79 indels associated with HRs that might otherwise be interpreted as potential errors if encountered in a pyrosequencing library.
Approximately 19% of the pyrosequences matched the near full-length PCR clones that comprised the different phylogroups (Figure 2). Of these, ~83–100% (depending on phylogroup) were associated with vent emissions, streamers or mixing zone samples (i.e., high temperature samples). For the balance of the pyrosequence reads (~81%), collector's curves were constructed to identify a conservative OTU clustering criterion. As expected, as OTU clustering criteria became more conservative total diversity estimates declined (Figure 3), with the collector's curve constructed for 96% identity suggesting that the pyrosequencing data captured a majority of the Nanoarchaeaota diversity in the lake sampling scheme that spanned north-south and east-west, as well as various hydrothermal features. Taking into account all habitat types (photic zone, vents, vent-associated streamers, and vent-lake water mixing zones) and examining the lake by region, most of the OTUs were found in the West Thumb and Inflated Plain regions of the lake, again corresponding to the lake floor hydrothermal vents, which are primarily found in these regions of the lake (Figure 1).
Figure 3.
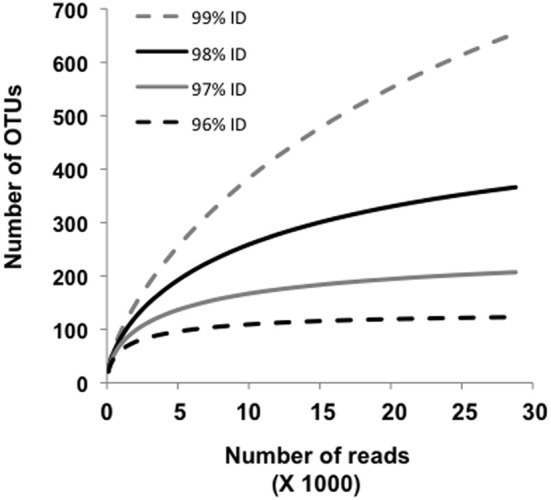
Collector's curves estimating the number of Nanoarchaeota OUTs identified for all samples, and as a function of sequence identity set at 96, 97, 98, and 99%. The curves depict the complete pyrosequencing data set after quality trimming according to Kunin et al. (2010) and OUT clustering as described by Huse et al. (2010).
Interestingly, even at the conservative OTU clustering employed, there was single, relatively small OTU (70 reads) that was unique to the photic zone sample taken in the Southeast Arm (Figure 1), a region of the lake where no known vent activity exists. This particular OTU was also exclusive to the largest size-class biomass (smaller than 20 μm but larger than 3.0 μm) and did not group with any of the major phylogroups identified using near full-length sequences (discussed above, Figure 2). The cohesiveness of this OTU was further examined by assessing the shared identity of the reads at higher levels of sequence identity. At 99%, this OTU disaggregated to single groups of 33 reads and 12 reads, four groups of four reads each, four groups of two reads each, and one singleton. At 98% identity, it broke into three groups of 49, 16, and 5 reads, whereas at 97% identity it remained complete at 70 reads.
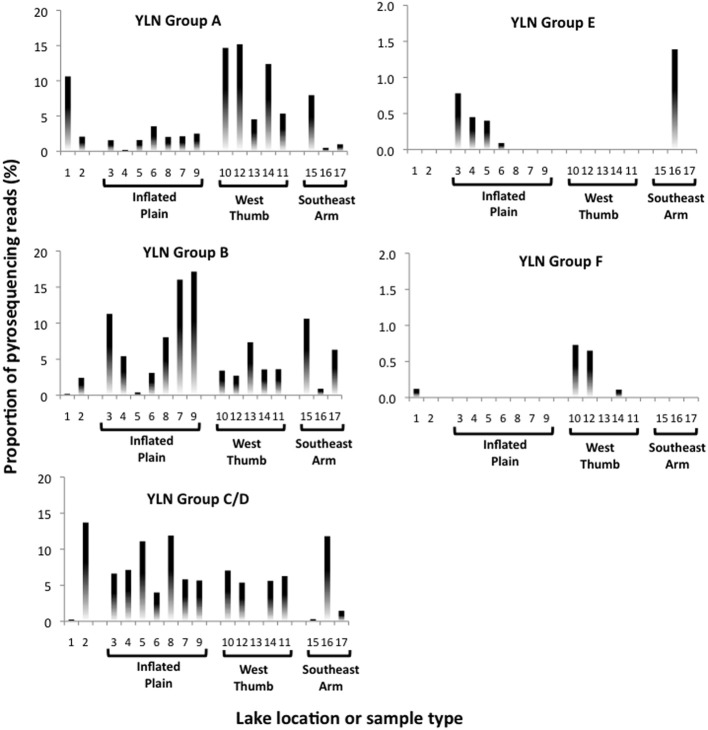
Photic zone water samples from the West Thumb region failed to generate a PCR product, suggesting that at least at the time of sampling the Nanoarchaeota were absent or below PCR detection in this portion of the lake. Other potentially interesting distribution patterns were revealed when examining pyrosequence distribution as matched to the phylogroups identified in the near full length clones (see Figure 2). Whereas phylogroups A, B, and C/D were found throughout the lake (Figure 4), phylogroups E and F signatures appeared to exhibit patterns. For example, phylogroup E was predominantly (90% of the group E matching pyroreads) found in photic zone samples in the Inflated Plain and in particular the Southeast Arm, but was not detectable in the West Thumb vents nor in the Elliot's Crater or Mary Bay vent emissions (Figure 4). By contrast, phylogroup F (F1 and F2 combined for analysis) was primarily (95%) found associated West Thumb vents, but was undetectable in any of the Inflated Plain vent or photic samples, nor in the Southeast Arm photic water samples (Figure 4).
Figure 4.
Nanoarchaeota phylogroup distribution across Yellowstone Lake based on 99% pyroread sequence match to near full length Sanger-sequenced clones shown in Figure 2. Data shown the proportional representation of each phlyogroup in the pyrosequence library generated for each site or sample. Location/sample numbers are keyed to Table 1, which provides a detailed description for each environment. Note the different y-axis scales.
Finally, in an attempt to gain more information about the Nanoarchaeota in this lake, fluorescent in situ hybridization (FISH) probes were designed, prepared, and applied to raw lake photic water samples. These FISH probing attempts were hampered by visual interference from small sized lake debris particles and presumably low target density. The latter was concluded from the weak Nanoarchaeota PCR amplicon strength in these samples relative to the much more robust Bacteria or Archaea 16S rRNA gene amplification products observed in previous studies on this lake (Clingenpeel et al., 2011; Kan et al., 2011).
Discussion
The Nanoarchaeaota are represented by a single co-cultured and characterized isolate, N. equitans (Huber et al., 2002). As such, it is unreasonable to assume that this lone isolate adequately represents this proposed phylum. Indeed, a recent single cell genomics study highlighted differences between N. equitans and Nanoarchaeaota cells captured from Obsidian Pool in Yellowstone (Podar et al., 2013). Initial views of the Nanoarchaeaota being hyperthermophiles associated with Ignicoccus have given way to subsequent molecular-based surveys that found the Nanoarchaeaota occurring in extreme environments where Ignicoccus is not known to reside (Casanueva et al., 2008; Podar et al., 2013). As environmental clone data has accumulated, biogeographical patterns have begun to emerge, which were considerably strengthened and expanded by the current study. The clone sequences acquired in this study firmly establishes the Yellowstone Nanoarchaeaota as a robust and distinct phylogenetica clade separate from those in geographically distant locations (Figure 2).
Indels were not the primary source of sequence diversity that defined the primary phylogroups observed in this lake (Figure 2), but their frequency was significant. Some indels in the pyrosequencing libraries were no doubt errors associated with HRs (Kunin et al., 2010), which are very dense in the Nanoarchaeota 16S gene sequences examined in this study. As averaged from six randomly selected near full-length clones, the frequency of HR per clone was: 3 bp HR = 73 ± 2; 4 bp HR = 21 ± 3; 5 bp HR = 11 ± 1; 6 bp HR ~1 ± 1; or 7 bp HR = 0.3 ± 0.6. As a contrast example, this is roughly two-fold that found across the longer (1542 bp) length of the seven 16S rRNA genes of Escherichia coli strain K12: 3 bp HR = 55 ± 2; 4 bp HR = 11 ± 2; 5 bp HR = 4 ± 0; 6 bp HR ~1 ± 0; or 7 bp HR = 0. Regardless of the HR issue, however, Sanger sequencing of clones from two different PCR libraries demonstrated that some of these indels appear real. This conclusion is based on multiX coverage for nucleotide assignments that appeared as an indel relative to other clone sequences and suggests the indels are a natural feature of the Nanoarchaeota 16S rRNA gene. For logistical and cost reasons, the absolute frequency of these indels would be very difficult to pinpoint.
Rarefaction analysis set at 96% identity suggested complete coverage of the lake Nanoarchaeota as defined by the pyrosequencing library (Figure 3). A small proportion (~2%) of the 96% identity defined OTUs were detected in all lake samples examined, suggesting some level of lake-wide mixing and is consistent with what was observed with pyroreads assigned to phylogroups A, B, and C/D (Figure 4) as well as what we have reported for the Bacteria (Clingenpeel et al., 2011) and Archaea (Kan et al., 2011) in this lake. And while the pyrosequences that matched all of the major phylogroups depicted in Figure 2 were primarily found associated with the lake floor hydrothermal features (Figure 4), there were instances such as with phylogroups E and F where abundance appeared to be biased toward photic zone waters (Figure 4). Further, the pyrosequencing libraries contained a single OTU that was not found in the high temperature samples, but rather only in the largest filtration size class and only in the Southeast Arm photic zone waters. Nanoarchaeota being associated with the lake floor hydrothermal vents was not necessarily unanticipated, but the potential for photic zone Nanoarchaeota was not expected. Occurrence of the Nanoarchaeota in low temperature environments has been documented previously (Casanueva et al., 2008), establishing a precedent for low temperature versions of this interesting microorganism. This will be the subject of follow-up work.
At this juncture, linking the Yellowstone Lake Nanoarchaeota to potential host phylotypes is not possible, except to conclude with near certainty that the potential host list does not include an Ignicoccus-like lineage. Our previous studies of this lake yielded several lines of evidence demonstrating freshwater parallels to important marine organisms; e.g., Prochloroccus (Clingenpeel et al., 2011) and a Nitrosopumilus-like archaean (Kan et al., 2011). However, an Ignicoccus-like lineage was notably absent in pyrosequencing surveys totaling 51,017 454-FLX reads (Kan et al., 2011) and 262,173 454-Titanium reads [Community Cyberinfrastructure for Advanced Microbial Ecology Research & Analysis (CAMERA); https://portal.camera.calit2.net/gridsphere/gridsphere?cid=microgenome]. Preliminary evidence of Nanoarchaeota associated (physically attached) with a Pyrobaculum-shaped bacterium (Stetter et al., 2005) is consistent with the view that other Archaea can serve as hosts. Further, a recent report by Podar et al. (2013) described a relationship between Nanoarchaeota from Obsidian Pool (YNP) and a Sulfolobales-like archaeon that co-isolated in cell sorting experiments used for single cell genome sequencing efforts. Non-Ignecoccus hosts would also seem the case for the Nanoarchaeota documented for non-thermal hypersaline mats (Casanueva et al., 2008).
In summary, this study revealed the very significant Nanoarchaeota 16S rRNA gene diversity occurring in natural populations associated with the hydrothermal vents in Yellowstone Lake as well as lineages that may reside in photic waters. Phylogenetically, these organisms form a clade that clusters with the YNP Obsidian Pool Nanoarchaeota clone that is distinctly separate from the N. equitans and the Kamchatka Nanoarchaeota. The hosts for the Yellowstone Lake Nanoarchaeota are unknown at present, but we conclude do not include Ignicoccus.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported primarily by a grant from the Gordon and Betty Moore Foundation (Grant No. 1555), with additional funding from the National Science Foundation (EPS-1101342), National Park Service Centennial Challenge Match Program (PMIS No. 137808) and the Office of Science of the U.S. Department of Energy (Contract No. DE-AC02-05CH11231). Any opinions, findings and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the Gordon & Betty Moore Foundation or the National Science Foundation. Work was conducted under NPS research permit No. 5700.
References
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Auchtung T. A., Takacs-Vesbach C. D., Cavanaugh C. M. (2006). 16S rRNA phylogenetic investigation of the candidate division “Korarchaeota.” Appl. Environ. Microbiol. 72, 5077–5082 10.1128/AEM.00052-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balistrieri L. S., Shanks W. C., III, Cuhel R. L., Aguilar C., Klump J. V. (2007). “The influence of sub-lacustrine hydrothermal vents on the geochemistry of Yellowstone Lake,” in Integrated Geoscience Studies in the Greater Yellowstone Area – Volcanic, Tectonic, and Hydrothermal Processes in the Yellowstone Geoecosystem, U.S. Geological Survey Professional Paper 1717, ed Morgan L. A. (Reston, Virginia: ), 173–199 [Google Scholar]
- Barns S. M., Fundyga R. E., Jeffries M. W., Pace N. R. (1994). Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment. Proc. Natl. Acad. Sci. U.S.A. 91, 1609–1613 10.1073/pnas.91.5.1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson N. G. (1961). Limnology of Yellowstone Lake in Relation to the Cutthroat Trout. Research Report 56. Washington, DC: US Fish and Wildlife Service, 33. [Google Scholar]
- Botero L. M., D'Imperio S., Burr M., McDermott T. R., Young M., Hassett D. J. (2005). Poly(A) polymerase modification and reverse transcriptase PCR amplification of environmental RNA. Appl. Environ. Microbiol. 71, 1267–1275 10.1128/AEM.71.3.1267-1275.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd E. S., Jackson R. A., Encarnacion G., Zahn J. A., Beard T., Leavitt W. D., et al. (2007). Isolation, characterization, and ecology of sulfur-respiring crenarchaea inhabiting acid-sulfate-chloride-containing geothermal springs in Yellowstone National Park. Appl. Environ. Microbiol. 73, 6669–6677 10.1128/AEM.01321-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanueva A., Galada N., Baker G. C., Grant W. D., Heaphy S., Jones B., et al. (2008). Nanoarchaeal 16S rRNA gene sequences are widely dispersed in hyperthermophilic and mesophilic halophilic environments. Extremophiles 12, 651–656 10.1007/s00792-008-0170-x [DOI] [PubMed] [Google Scholar]
- Clingenpeel S., Macur R. E., Kan J., Inskeep W. P., Lovalvo D., Varley J., et al. (2011). Yellowstone Lake: high-energy geochemistry and rich bacterial diversity. Environ. Microbiol. 13, 2172–2185 10.1111/j.1462-2920.2011.02466.x [DOI] [PubMed] [Google Scholar]
- Cole J. R., Wang Q., Cardenas E., Fish J., Chai B., Farris R. J., et al. (2009). The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37, D141–D145 10.1093/nar/gkn879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis D. G., Bizzoco R. W., Kelley S. T. (2008). Halophilic Archaea determined from geothermal steam vent aerosols. Environ. Microbiol. 10, 1582–1590 10.1111/j.1462-2920.2008.01574.x [DOI] [PubMed] [Google Scholar]
- Flores G. E., Campbell J. H., Kirshtein J. D., Meneghin J., Podar M., Steinberg J. I., et al. (2011). Microbial community structure of hydrothermal deposits from geochemically different vent fields along the Mid-Atlantic Ridge. Environ. Microbiol. 13, 2158–2171 10.1111/j.1462-2920.2011.02463.x [DOI] [PubMed] [Google Scholar]
- Flores G. E., Shakya M., Meneghin J., Yang Z. K., Seewald J. S., Geoff Wheat C., et al. (2012). Inter-field variability in the microbial communities of hydrothermal vent deposits from a back-arc basin. Geobiology 10, 333–346 10.1111/j.1472-4669.2012.00325.x [DOI] [PubMed] [Google Scholar]
- Fouke B. W., Farmer J. D., Des Marais D. J., Pratt L., Sturchio N. C., Burns P. C., et al. (2000). Depositional facies and aqueous-solid geochemistry of travertine-depositing hot springs (Angel Terrace, Mammoth Hot Springs, Yellowstone National Park, U.S.A.). J. Sediment. Res. A Sediment. Petrol. Process. 70, 565–585 10.1306/2DC40929-0E47-11D7-8643000102C1865D [DOI] [PubMed] [Google Scholar]
- Hohn M. J., Hedlund B. P., Huber H. (2002). Detection of 16S rDNA sequences representing the novel phylum “Nanoarchaeota”: indication for a wide distribution in high temperature biotopes. Syst. Appl. Microbiol. 25, 551–554 10.1078/07232020260517698 [DOI] [PubMed] [Google Scholar]
- Huber H., Hohn M. J., Rachel R., Fuchs T., Wimmer V. C., Stetter K. O. (2002). A new phylum of Archaea represented by a nano-sized hyperthermophilic symbiont. Nature 417, 63–67 10.1038/417063a [DOI] [PubMed] [Google Scholar]
- Hugenholtz P., Pitulle C., Hershberger K. L., Pace N. R. (1998). Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180, 366–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse S. M., Welch D. M., Morrison H. G., Sogin M. L. (2010). Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ. Microbiol. 12, 1889–1898 10.1111/j.1462-2920.2010.02193.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inskeep W. P., Rusch D. B., Jay Z. J., Herrgard M. J., Kozubal M. A., Richardson T. H., et al. (2010). Metagenomes from high-temperature chemotrophic systems reveal geochemical controls on microbial community structure and function. PLoS ONE 5:e9773 10.1371/journal.pone.0009773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson C. R., Langner H. W., Donahoe-Christiansen J., Inskeep W. P., McDermott T. R. (2001). Molecular analysis of microbial community structure in an arsenite-oxidizing acidic thermal spring. Environ. Microbiol. 3, 532–542 10.1046/j.1462-2920.2001.00221.x [DOI] [PubMed] [Google Scholar]
- Kan J., Clingenpeel S., Macur R. E., Inskeep W. P., Lovalvo D., Varley J., et al. (2011). Archaea in Yellowstone Lake. ISME J. 5, 1784–1795 10.1038/ismej.2011.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karavaĭko G. I., Golyshina O. V., Troitskii A. V., Val'ekho-Roman K. M., Golovacheva R. S., Pivovarova T. A. (1994). Sulfurococcus yellowstonii sp. nov–a new species of iron- and sulfur-oxidizing thermoacidophilic Archaeobacterium. Mikrobiologiia 63, 668–682 [PubMed] [Google Scholar]
- Kunin V., Engelbrektson A., Ochman H., Hugenholtz P. (2010). Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ. Microbiol. 12, 118–123 10.1111/j.1462-2920.2009.02051.x [DOI] [PubMed] [Google Scholar]
- Lovalvo D., Clingenpeel S. R., McGinnis S., Macur R. E., Varley J. D., Inskeep W. P., et al. (2010). A geothermal-linked biological oasis in Yellowstone Lake, Yellowstone National Park, Wyoming. Geobiology 8, 327–336 10.1111/j.1472-4669.2010.00244.x [DOI] [PubMed] [Google Scholar]
- Ludwig W., Strunk O., Westram R., Richter L., Meier H., Kumar Y., et al. (2004). ARB: a software environment for sequence data. Nucleic Acids Res. 32, 1363–1371 10.1093/nar/gkh293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maidak B. L., Olsen G. J., Larsen N., Overbeek R., McCaughey M. J., Woese C. R. (1997). The RDP (Ribosomal Database Project). Nucleic Acids Res. 25, 109–111 10.1093/nar/25.1.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCliment E. A., Voglesonger K. M., O'Day P. A., Dunn E. E., Holloway J. R., Cary S. C. (2006). Colonization of nascent, deep-sea hydrothermal vents by a novel archaeal and nanoarchaeal assemblage. Environ. Microbiol. 8, 114–125 10.1111/j.1462-2920.2005.00874.x [DOI] [PubMed] [Google Scholar]
- Morgan L. A., Blackwell D. D., Spafford R. E., Smith R. B. (1977). Heat flow measurements in Yellowstone Lake and the thermal structure of the Yellowstone caldera. J. Geophys. Res. 82, 3719–3732 10.1029/JB082i026p03719 [DOI] [Google Scholar]
- Morgan L. A., Shanks W. C., III, Lovalvo D., Johnson S. Y., Stephenson W., et al. (2003). Exploration and discovery in Yellowstone Lake: results from high-resolution sonar imaging, seismic reflection profiling, and submersible studies. J. Volcanol. Geotherm. Res. 122, 221–242 10.1016/S0377-0273(02)00503-6 [DOI] [Google Scholar]
- Morgan L. A., Shanks W. C., III, Pierce K. L., Lovalvo D. A., Lee G. K., Webring M. W., et al. (2007). “The floor of Yellowstone Lake is anything but quiet – new discoveries from high resolution sonar imaging, seismic-reflection profiling, and submersible studies,” in Integrated Geoscience Studies in the Greater Yellowstone Area – Volcanic, Tectonic, and Hydrothermal Processes in the Yellowstone Geoecosystem, ed Morgan L. A. (Reston, Virginia: U.S. Geological Survey Professional Paper 1717; ), 95–126 [Google Scholar]
- Podar M., Makarova K. S., Graham D. E., Wolf Y. I., Koonin E. V., Reysenbach A. L. (2013). Insights into archaeal evolution and symbiosis from the genomes of a nanoarchaeon and its inferred crenarchaeal host from Obsidian Pool, Yellowstone National Park. Biol. Direct 8:9 10.1186/1745-6150-8-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E., Quast C., Knittel K., Fuchs B., Ludwig W., Peplies J., et al. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35, 7188–7196 10.1093/nar/gkm864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reysenbach A. L., Ehringer M., Hershberger K. (2000). Microbial diversity at 83 degrees C in Calcite Springs, Yellowstone National Park: another environment where the Aquificales and “Korarchaeota” coexist. Extremophiles 4, 61–67 [DOI] [PubMed] [Google Scholar]
- Roussel E. G., Konn C., Charlou J. L., Donval J. P., Fouquet Y., Querellou J., et al. (2011). Comparison of microbial communities associated with three Atlantic ultramafic hydrothermal systems. FEMS Microbiol. Ecol. 77, 647–665 10.1111/j.1574-6941.2011.01161.x [DOI] [PubMed] [Google Scholar]
- Rusch D. B., Halpern A. L., Sutton G., Heidelberg K. B., Williamson S., Yooseph S., et al. (2007). The Sorcerer II global ocean sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 5:e77 10.1371/journal.pbio.0050077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage S. L., Lawrence R. L., Custer S. G., Jewett J. T., Powell S. L., Shaw J. A. (2012). Analyzing change in Yellowstone's terrestrial emittance with Landsat imagery. GISci. Remote Sens. 49, 317–345 10.2747/1548-1603.49.3.317 [DOI] [Google Scholar]
- Schloss P. D., Westcott S. L., Ryabin T., Hall J. R., Hartmann M., Hollister E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear J. R., Walker J. J., McCollom T. M., Pace N. R. (2005). Hydrogen and bioenergetics in the Yellowstone geothermal ecosystem. Proc. Natl. Acad. Sci. U.S.A. 102, 2555–2560 10.1073/pnas.0409574102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetter K. O., Hohn M. J., Huber H., Rachel R., Mathur E., Hedlund B., et al. (2005). “A novel 478 kingdom of parasitic Archaea,” in Geothermal Biology and Geochemistry in Yellowstone 479 National Park. Proceeding of the Thermal Biology Institute Workshop, Yellow-stone National 480 Park, WY, October 2003, eds Inskeep W. P., McDermott T. R. (Bozeman, MT: Montana State University Publications; ), 249–259 [Google Scholar]
- Wang Q. G., Garrity M., Tiedje J. M., Cole J. R. (2007). Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters E., Hohn M. J., Ahel I., Graham D. E., Adams M. D., Barnstead M., et al. (2003). The genome of Nanoarchaeum equitans: insights into early archaeal evolution and derived parasitism. Proc. Natl. Acad. Sci. U.S.A. 100, 12984–12988 10.1073/pnas.1735403100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T., Lyons S., Aguilar C., Cuhel R., Teske A. (2011). Microbial communities and chemosynthesis in Yellowstone Lake sublacustrine hydrothermal vent waters. Front. Microbiol. 2:130 10.3389/fmicb.2011.00130 [DOI] [PMC free article] [PubMed] [Google Scholar]