Figure 5.
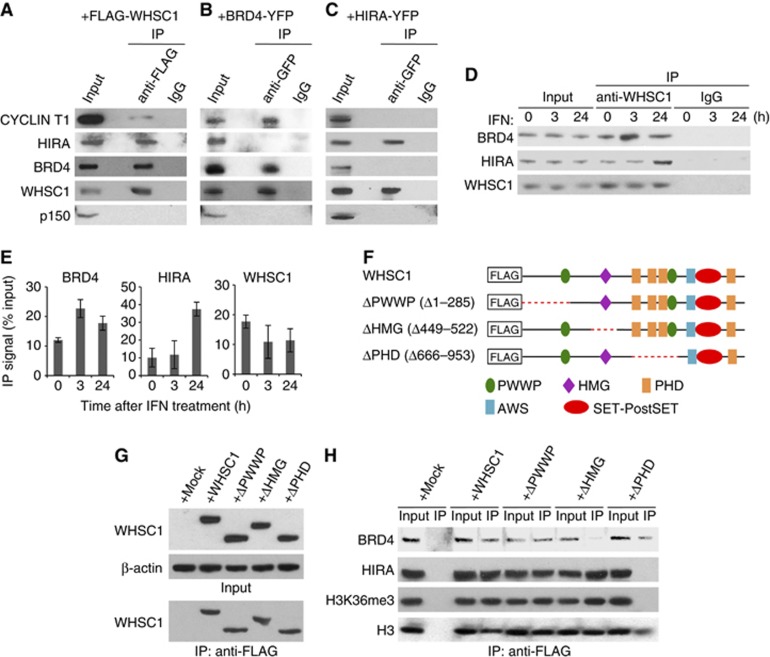
WHSC1 interacts with BRD4 and HIRA independently. (A) Whole cell extracts from Whsc1−/− cells transiently expressing FLAG-WHSC1 were immunoprecipitated with anti-FLAG antibody, and tested for co-precipitation of indicated proteins by immunoblotting. Normal IgG was used as a control. Input represents 10% of total protein. (B) Extracts from NIH3T3 cells expressing BRD4-YFP were immunoprecipitated with anti-GFP antibody, and tested for co-precipitation of indicated proteins by immunoblotting. (C) Whole cell extracts from WT cells transiently expressing HIRA-YFP were immunoprecipitated with anti-GFP antibody, and tested for co-precipitation of indicated proteins by immunoblotting. (D) Whole cell extracts from WT cells treated with IFN for indicated times were immunoprecipitated with anti-WHSC1 antibody, and tested for co-precipitation of BRD4 and HIRA. (E) Graphic representation of the experiments shown in (D). The amounts of precipitated proteins were quantified and normalized by input protein. Values are the average of two assays ±s.d. (F) A diagram of FLAG-tagged WHSC1 deletion mutants. The red dots indicate deleted regions. (G) Whole cell extracts from Whsc1−/− cells transiently expressing above deletion constructs were immunoblotted with anti-WHSC1 antibody (left) or immunoprecipitated with anti-FLAG antibody, followed by immunoblotting with anti-WHSC1 antibody (right). (H) Proteins immunoprecipitated from above cells with anti-FLAG antibody were immunoblotted for BRD4, HIRA, H3K36me3, or total H3.
Source data for this figure is available on the online supplementary information page.