FIG. 6. RNase H assay consisted of 5’ 32P end labeled 38-mer RNA annealed to a 32-nt unlabeled DNA primer.
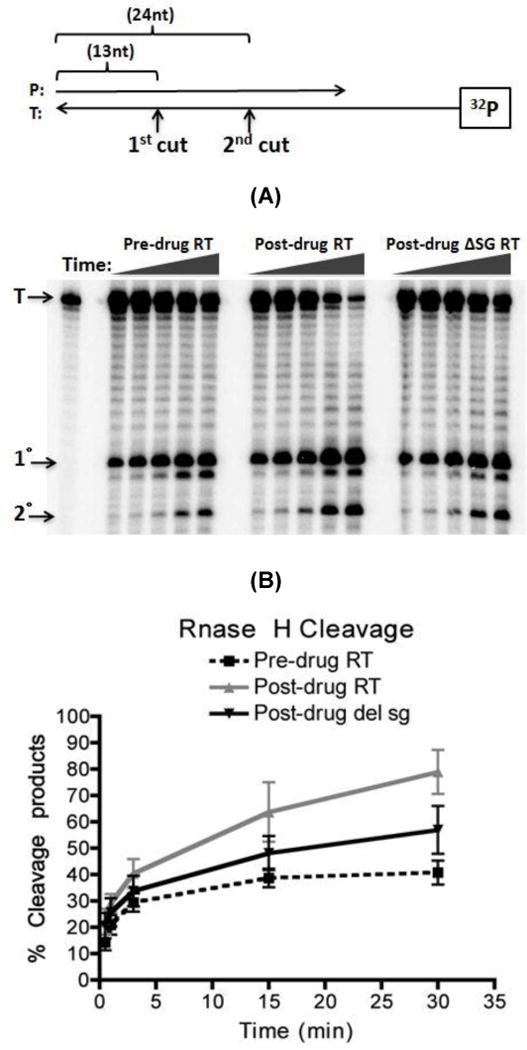
(A) The same amount of RT activities was used as in Fig. 3 for this assay. The reaction was carried out in a time course assay (0.5, 1, 3, 15, 30 minutes) in the absence of dNTP and quenched with the addition of EDTA. RNase H cleavages yielded a 24 nt primary cut product and a 13 nt secondary cut product. (B) Quantification of RNase H 5’ 32P end labeled 38-mer RNA degradation from Fig. 6A shows percent of degraded RNA over time. Percent degraded was calculated base on amount of (saturated volume from 1 □ + 2 □ cleavages)/(saturated volume from 1 □ + 2 □ + Uncut) × 100
