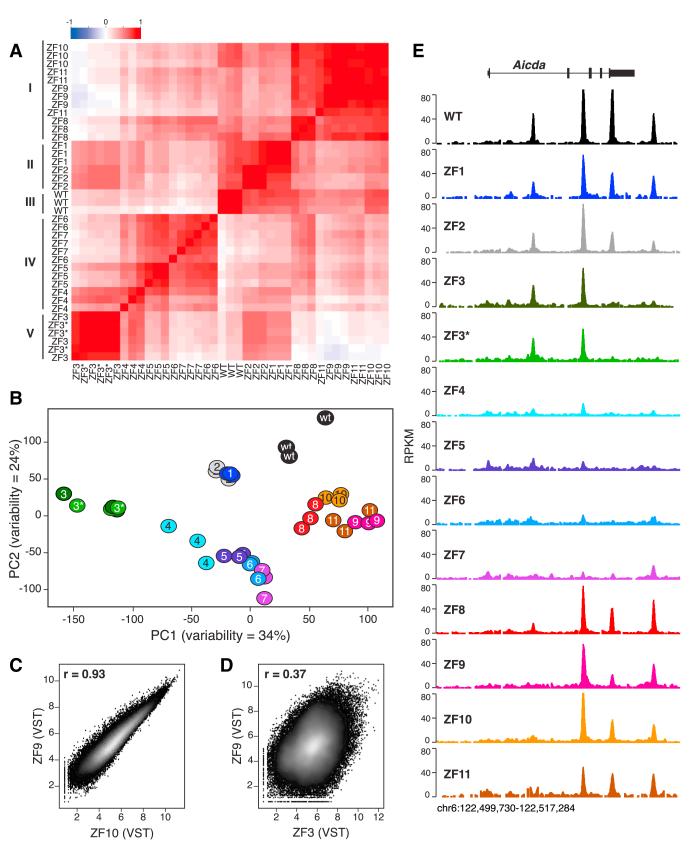
Figure 3. CTCF ZFs Cluster into DNA Binding Subdomains.
(A) Pearson’s correlation matrix analysis of variance-stabilized CTCF ChIP-seq data at 14,804 sites that showed significant changes in CTCF binding. Five distinct clusters (ZF8–11, ZF1–2, WT, ZF4-7, and ZF3–3*) are highlighted. Scale represents Pearson’s r (from ~1 to 1).
(B) Principal component analysis of CTCF ChIP-seq data sets.
(C) Scatterplot comparison of variance-stabilizing transformed (VST) ChIP-seq data between ZF9 and ZF10 CTCF mutants. Correlation is provided via Pearson’s coefficient r.
(D) Same comparison as (C) between ZF9 and ZF3.
(E) CTCF WT and mutant binding profiles at mouse Aicda locus in chromosome 6. ChIP-seq samples were normalized as RPKM. See also Figure S3.